White Paper
Torrent Variant Detection Algorithms (3.4)
This white paper describes algorithms developed byIon Torrent for the detection of SNPs and small indels on the Ion Proton andIon PGM platforms. There are three core algorithms: one for SNP detection, one for detection of insertions and deletions (GATK-based), and one for long indels (assembly).
SNP Detection
The SNP Detection algorithm takes as input a BAM file of aligned reads (for example, a BAM file produced by TMAP, the Torrent aligner), and tiles the genome (or subset of the genome defined by a BED file), evaluating evidence for a variant by looking at a pileup of the reads.
Our SNP detection algorithm includes filters by read, base, frequency, and genome position, and it includes statistical modeling of data that passes the filters to evaluate the probability that a SNP exists at a candidate genome position. If the position passes the filters and there is sufficient evidence, it is called as a SNP and assigned a genotype and a p-value.
Two methods based on statistical modeling are implemented for base-space SNP prediction: Bayesian and frequentist. The Bayesian methodis currently only implemented for germ-line SNP detection and is most useful when coverage is low to medium (between 10-50). The frequentist approach is used for positions with higher coverage and can be used to detect low frequency variants for example, it is used by default for positions with filtered coverage greater than 60x.
Some common filtering occurs before the algorithm. Common filters are the following:
- A read-level filter R emoves reads with low mapping QV.
- A position-level filter C hecks each position of the reference to see if there are enough reads mapped to an alternative allele. This filter is controlled by allele frequency (a user-defined parameter).
- A position-level filter R emoves positions where the reads for the variants have significantly more strand bias than the reads covering the position.
- A base-level filter R emoves bases of reads with poor base QV.
Extreme strand bias in reads predicting a variant often results in false positive variant calls. To avoid these, we use a method called relative strand bias filtering. For a given genomic position, let Cp and Cm be the numbers of reads in the plus and minus strands that cover this position, and let Vp and Vm be the numbers of reads in plus and minus strands that match the variant. The relative variant strand bias is defined by:
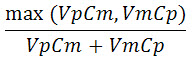
Positions in which a potential variant has a high relative strand bias are filtered out. The relative bias is found by first normalizing strand count for the variant based on the bias of the total set of reads at the position, then calculating the strand bias on the normalized count.
Hotspot positions in a sequencing sample are much more likely to have mutations. In human, while the background chance of SNP at any random position is about 1 in 200 or more, a hotspot position can be 10s or even 100s times more likely to have a SNP. Our normal filters and p-value cutoffs for SNP calling are overly conservative for a hotspot position. In order to detect hotspot SNPs, we allow a more aggressive set of filter settings and p-value cutoff for hotspot positions.
Once a position with a potential variant passes the filter and reads covering the position are collected, we choose one of the two algorithms to determine the genome type and p-value(s) of prediction.
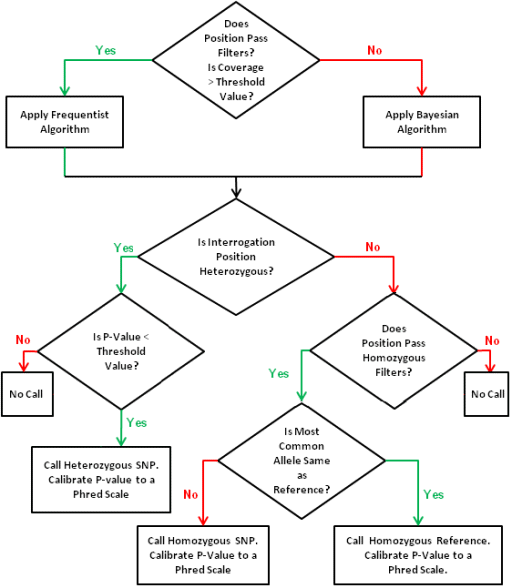
Bayesian Method
When coverage is below a threshold (for example, 60x), we use the Bayesian method. From the set of reads covering the position, we have the number of reads supporting each of thefour alleles. We use the most common allele, mca, and second most common allele, sca . The Bayesian algorithm checks threehypotheses: No SNP (ref), homozygous SNP (if mca is not a reference allele), and heterozygous SNP (mca/sca), by calculating P(ref|R) , P(Het|R) , and P(Hom|R) , where R is the set of the reads, and each of the posterior probabilities is calculated using Bayes' theorem:
P(H|R)=P(R|H)P(H)/P(R) , where P(H) is the prior and P(R) is a constant, and P(R|H) is easily calculated using the base QV to estimate the chance that a base is correct.
If P=P(ref|R)+P(Hom|R)+P(Het|R) , then the likelihood of each prediction is P(H|R)/P , and the hypothesis with the highest likelihood is called.
Frequentist Method
When coverage is high, we can use the frequentist method, whicheffectively estimates P(ref|R) . If the genome position does not have an alternative allele (i.e., it has no SNP), we dothis by estimating the expected number of reads matched to an alternative allele. Using the QV of all reads, we estimate the error rate of the bases covering this position. This error rate times the coverage depthis the number of non-reference reads expected. From the set of reads, we know the actual number of reads matching the alternative allele. A Poisson distribution is used to estimate the likelihoodof this happeningby chance. Low-frequency variants can be detected by this method. The variants are reported in a VCF file.
SNPs called near homopolymer (HP) positions tend to be more likely to be false positives because of a higher chance of mismapping, in addition to the higher error rate at these positions. Also, longer HPs are more likely to cause false positives. We model this observation by adding an HP penalty. Formally, for any HP of length L we define a penalty function P(L)=a+bL when L<C and P(L)=a+bL+d otherwise, where a , b , c , d are user-specified parameters. In the frequentist method, the error rate of each base was determined by its base QV, and for any base belong to a HP of length L , we multiply P(L) by the error rate calculated from the base QV. The process is also equivalent to lowering the base QV of HP by a certain amount determined by length.
Indel Detection - GATK
Overall Ion's Unified Genotyper in the current release makes use of not only the flow order and flow intensities, but also Ion's base calls, CIGAR alignment, and base quality calls.
The indel caller in the Torrent Variant Caller (TVC) implements four consecutive steps. The first two steps are designed to accurately call short indels of less than 15bp. The third step is designed to reconstruct long indels 1570bp in size. The fourth step filters candidate indels and produces final calls.
First step
In the first step, we traverse all genomic positions limited to the regions provided in the BED file. For each position, we generate a list of observed indels by inspecting a pileup of base-space alignments crossing that position. By default, each indel that is present at a frequency of 10% or higher is included in the set of candidate variants. This step is implemented by calling GATK UnifiedGenotyper in the multi-threaded mode with a certain level of downsampling for speed. The list of candidate indels contains most of the true indels and a large number of false indels caused by misalignments and sequencing errors.
Second step
In general in this step, each candidate indel is rescored using a Bayesian framework that estimates significance of the indel from flow intensities and flow-space alignments across reads that have the indel in their base-space sequence. This step is implemented by calling a modified version of the GATK UnifiedGenotyper that has two major additional functionalities:
a. Extracts flow signal from BAM. Realigns flow signal to base calls. Collects flow intensities from reads supporting an indel. Calculates a new Bayesian score (with calls external to GATK).
b. Correctly calculates the number of reads on each strand that support the reference allele or indel variant.
All the candidate indels identified in step 1 are classified into one of the following three categories:
- Indels in a homopolymer region (example: 'TTTT' -> 'TTT')
- Indels within a multi-nucleotide repeat region (MNR), where one or more copies of a series of 2-, 3-, or 4-nucleotide bases are either inserted or deleted(example: 'ATATAT' -> 'ATAT')
- Other indels of one or more bases that are not in a homopolymer or multi- nucleotide repeat region
Previous releases refer to the MNR category as di-nucleotide and tri-nucleotide repeats. 3.x releases also consider4-nucleotide repeats in this category.
Each candidate category receives a different approach, as described in the following sections.
Indels in a homopolymer region
The candidate indels classified as homopolymer indels are further evaluated using a separate module called Flow Peak Estimator (FPE). FPE develops the distribution of flow signals from all the reads spanning the candidate position and does a flow-space evaluation using the flow signal distribution. The module assigns a score to all candidate indels and filters out all candidates below a score threshold.
The FPE module fits the flow signal distribution to a Gaussian model and uses a least-squares min approach to determine the number of peaks in the flow signal distribution.The presence of two signal peaks, one centered around the reference homopolymer length and the other centered around the candidate homopolymer length, is used as evidence for a heterozygous sample. The presence of a single peak centered around candidate homopolymer length is evidence of a homozygous indel. Several quantitative measures are used to identify error modes in the flow signal distribution.
Strand-bias is the principal error mode that results in noisy bi-modal flow signal distribution (where only one strand contributes to signal peak around candidate homopolymer length). A k-means clustering approach is used to identify the two clusters (if any) within all the flow signals contributing to the candidate indel position. The strand-bias estimate is calculated from the number of flow signals contributing to the two identified clusters from the postive and negative strands.The strand-bias threshold is controlled by the parameter
hp_stb_max_two_peaks_relative_bias
, which is set to default value of 0.8. The minimum coverage required to calculate this flow space based strand-bias estimate is controlled by the parameter
hp_stb_fpe_min_coverage
and the default value is set to 50.
If the coverage at any candidate indel position is below this threshold, then the position is no longer consider as an MNR candidate and is evaluated as a potential indel of one or non-consecutive bases:
- The indel is scored by the Bayesian framework
- The strand-bias is calculated using base space (number of reference and candidate alleles from each strand).
The standard deviation of the flow signal distribution is used as a measure to identify noisy signals at any given candidate indel position (homozygous variants in particular). The threshold for standard deviation of uni-modal flow signal distributions is controlled jointly by these parameters:
-
hp_max_single_peak_std_23Signifies the maximum allowed standard deviation for homopolymers of length up to 3 -
hp_max_single_peak_std_incrementSignifies the maximum allowed increment in standard deviation for homopolymers greater than length 3.
In the case of homozygous variants, the distribution of flow signals is expected to be uni-modal with the peak ideally centered around the homopolymer length of the variant allele (for example, around 600 for a 6mer
homopolymer
). The distance (
D
) between the observed and the expected mean of the flow signal peaks is another good quantitative measure of noise in flow signals and is controlled using the parameter
hp-max_peak_deviation
. A large
D
is an indicator of an error prone position and increasing this threshold would lower the specificity of the variants reported.
The allele frequencies, in case of heterozygous variants, is estimated using the flow signal distribution by taking the ratio of the areas under the two peaks in a bi-modal distribution (bi-modality is expected in flow signal distributions at heterozygous positions), rather than by simple counting of number reads supporting the each allele in base space.
hp_max_length parameter
This parameter, new in release 3.2, sets the maximum indel homopolymer length. HP indels longer than this value are not called. The default for this parameter varies by workflow:
- Ion AmpliSeq workflows 8
- Ion TargetSeq workflows 7
- Whole Genome workflow 7
Indels in a multi-nucleotide region
Candidate indels classified to be within a multi-nucleotide repeat region are further evaluated in the second step using a separate MNR module. This module again operates within the flow space rather than the base space to evaluate the candidate variants. The observed flow signals within the flow order spanning the candidate indel position is evaluated against the expected flow signals from both the null hypothesis (reference) and the hypothesis that variant is present. The measure of similarity between the observed and expected flow signals is used to generate a quantitative score for each candidate variant which is used in the final step of filter variants.
Other indels
All candidate indels that have not been classified as either of the other two types (in a homopolyer or in a multi-nucleotide repeat region) are further evaluated using a Bayesian framework that estimates significance of the indel from flow intensities and flow-space alignmentsacross reads that have the indel in their base-space sequence.
Second step output
The output of this step is a new VCF file that includes each candidate indel generated in the first step with the addition of the Bayesian score and strand counts in the info field of each variant.
Third step
In the third step, we sequentially traverse all reads in the BAM file and detect genomic regions with a large number of partially aligned reads that have >10bp of soft-clipped (non-aligned) sequence. This signals the possibility of presence of a large indel sequence that does not align to the reference. De novo assembly proceeds with soft-clipped sequences from partially aligned reads in a region and calls an indel if assembled sequence anchors to the reference. This step is designed to call long indels that are missed by the first two steps due to being soft-clipped by the aligner.
Fourth step
The fourth step step merges results from step 2 and 3 and filters them based on score, frequency, and relative strand bias. This step also sorts and produces the final list of variant calls.
Workflows
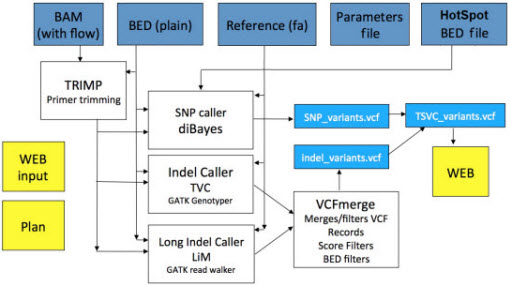
Torrent Variant Caller Workflow
-
First, in the Plan > Template tab, the user selects or creates a template appropriate to his or her sequencing experiment. (The template contains sequencing instructions, such as the sequencing application (
Ion AmpliSeq sequencing,
Whole Genome, TargetSeq, etc.),
reference, target region BED file (
f not using the whole genome application), and other options
.)
-
In the template, the user defines one of six workflows consisting of three possible region sizes (Whole Genome, TargetSeq, and Ion AmpliSeq), each of which may be called attwo frequencies (germ-line, somatic).
-
Once sequencing is performed,
TMAP
maps the reads to the reference, followed by a conversion to a flowspace alignment, and barcode splitting.
-
Reads are trimmed of primers for more accurate variant calling (because primers are not from the sample).
-
Summary reportsof coverage, variants, chromosomes, and HotSpots are generated.
-
The individual SNP and indel detectors are run, followed by a Bayesian Rescorer, which employs flowspace realignment, and filtering by parameters specific to the application.
- HotSpot variants are annotated and left-aligned, and a report is generated containing the summary, variant, and HotSpots tables, along with links to files that may be of interest to the user (BAM files, VCF files for SNPs and Indels, target region BED files).
Thevariant callingalgorithms are separately optimized for three workflows:
- Ion AmpliSeq Workflow, which includes the Ion AmpliSeq cancer research panels and custom Ion AmpliSeq panels.
- TargetSeq Workflow, with parameters optimized for hybridization-based target enrichment ("TargetSeq").
- Full Genome Workflow, which does not assume enrichment and does not require a target regions file.
These methods can be extended to the Ion TargetSeq Workflow and to verification of exome data. Previously we have extensively sequenced the HuRef exome, and reliably know where true positive variants are located.
Ion AmpliSeq Workflow
We report only variants within the target region (defined by the target BED file).
One specific module for the Ion AmpliSeq workflow is Primer Trimming. For AmpliSeq, we assign each read to the amplicon to which it most likely belongs and trim away any primer sequence, leaving only the insert sequence. This allows us to elegantly manage overlapping amplicons.
Several methods are used to verify indels. We obtained and sequenced 15 samples from the HapMap project, for which sequencing and variant data from the 1000 Genomes Project is publicly available.This provides us with, in principal, true positives for comparison, although they are somewhat limited in number per sample. To increase the absolute number of true positives, a dataset of simulated indels was generated by mapping reads from a HapMap sample to a modified hg19 reference. The strength of this method is that there are many true positives that are definitively known; however, simulation of this type is limited to homozygous calls.Early-access customer feedback is also a valuable source of information for verifying indels, especially when attempting to discover the reasons behind false positives.In some cases, customers have Sanger sequenced variants, providing us with the ability to examine known false negatives.
TargetSeq Workflow
We report only variants within the target region (defined by the target BED file).
Whole Genome Workflow
We report variants across the entire sequenced region.
However, the use of a target region BED file results in significant time savings. Without a regions of interest BED file, your plugin run may time out and fail to report results.

