References Management Guide
Torrent Suite Software space on Ion Community
References Management Guide TOC
Manage Target Regions Files and Hotspot Files
This page describes how to add, download, and remove target regions files and hotspot files.
Overview
Browser Extensible Data (BED) files and Variant Call Format (VCF) files supply chromosome positions or regions. When applied to a reference genome in the Torrent Browser, these files perform these two functions:
- Targeted regions of interest Specifies your regions of interest, for instance the amplified regions that are used with targeted sequencing. The complete Torrent Suite Software a nalysis pipeline, including plugins, is restricted to only the specified regions. (BED file only)
- Hotspot Instructs the Variant Caller to include these positions in its output files, including evidence for a variant and the filtering thresholds that disqualified a variant candidate. A hotspots file affects only the variantCaller plugin, not other parts of the a nalysis pipeline. (Either a BED or VCF file)
With the Torrent Browser, you add BED and VCF files to an existing reference. The reference must be listed in the Torrent Browser Admin > References tab before you can upload our BED or VCF files.
Your uploaded BED and VCF files are then available as an optionwhen you create a new template or planned run in the Plan tab. In the template and planned run wizard, menus on the Reference chevron page offer the BED and VCF files that you uploaded to a reference.
You can optionally upload multiple BED and VCF files to a reference.In the template and planned run wizard, you specify the BED or VCF files used for each template or each run.
Notes about hotspot files:
- By default the variantCaller plugin calls variant candidates at hotspot positions with more sensitivity than candidates at other positions. You can customize certain variantCaller parameters separately for hotspot candidates.
- The Torrent Browser also accepts VCF files as hotspot files.
Summary of steps to add a target regions or hotspots file
Before your analysis run or run registration (on the Planning page), you can add BED or VCF files to your genome reference:
- You use the Torrent Browser to upload the BED or VCF file from your local client machine to Torrent Suite Software.
- During file upload, the Torrent Browser verifies the BED or VCF file, and ensures that the BED or VCF file's coordinate regions are valid for the genome reference.
- The new BED or VCF file is then available as an option when you create a new run registration in the Planning tab. Your new file also appears in the Target Regions or HotSpots menus in the template and planned run wizard References chevron.
Modify a BED file
You can optionally modify a BED file before adding it to your reference genome. You can use this technique to avoid regions for which you do not want variants called (even if the variants appear in your sample).
You can modify a BED file only before uploading the file with the Torrent Browser.
Follow these instructions to modify a BED file:
-
Make a copy of your BED file. Rename the two files in a way that reflects changes you make to the regions being analyzed.
-
Open the BED file with a text editor.
-
Delete the lines for regions you do not want.
- Save the file.
If the region (or regions) appear in both your targeted regions BED file and in your hotspots BED or VCF file, you must delete the line for those regions from both types of BED file.
Supported file types
- Targeted regions of interest BED file only. Supported file extensions are .bed, .zip, and bed.gz.
- Hotspot BED file or VCF file. Supported file extensions are .bed, .vcf.gz, .zip, bed.gz, and .vcf.gz.
Upload a BED or VCF file
These instructions upload a BED or VCF file from your local client machine to Torrent Suite Software. These instructions apply to both targeted regions of interest files and hotspot regionsfiles.
You have the responsibility to avoid the following mismatch errors. The uploader does not always detect these errors:
- Upload a BED or VCF file to a reference genome of a different version (for example, an hg18 BED or VCF file with an hg19 reference).
- Upload a BED or VCF file for a different species.
- Upload a hotspots BED file as a targeted regions BED file, or upload a targeted regions BED file asa hotspots BED file.
Follow these steps to upload a target regions BED file or hotspots BED or VCF file to a reference:
-
In the
Reference
tab, click either the Hotspots or Target Regions tab in the left navigation panel:

The Hotspots (or Target Regions) page opens: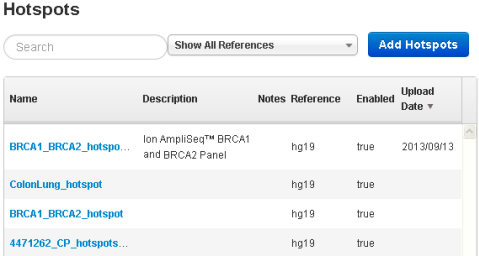
-
Click the
Add Hotspots
(or
Add Target Regions
) button in the top right corner.
The New Hotspots (or New Target Regions) page opens: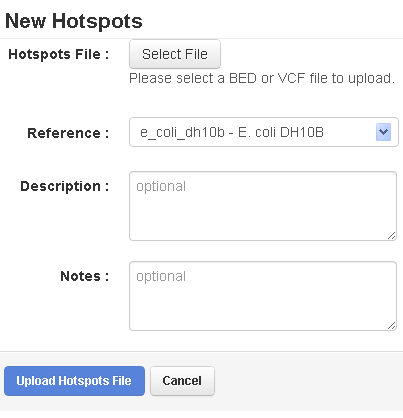
- Click the Select File button and browse to the file to be uploaded.
- In the Reference menu, be careful to select the correct reference. The new file can only be used with this reference.
- Add the optional (but recommended) description and notes.
- Click the Upload Hotspots File (or Upload Target Regions File ) button.
-
Wait while the file is validated:
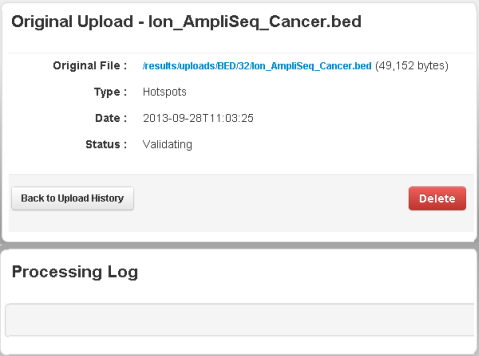
For large files, verification can take a couple minutes. Refresh your browser to check that validation is complete.
After upload
After validation, the Torrent Browser opens to the Hotspots detail page for your new file:
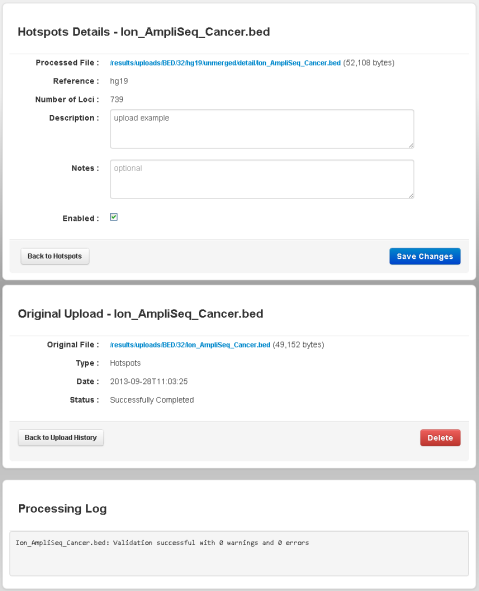
From this page, you can download the hotspots file or target regions file, remove the file from the system, and view the validation log.
Upload errors
Validation errors appear in the Processing Log section of the details page.
Some types of error do not appear in the Processing Log section. There are major problems that prevent validation from being attempted:
- Incorrect file format
- Incorrect file extension
- Zip contains 0 or multiple files
- A corrupted .zip .gz file
Download a hotspots or target regions file
Follow these steps to download a hotspots BED or VCF file, or a target regions BED file:
-
Go to the admin References tab and click either the Hotspots or Target Regions tab in the left navigation panel:

-
In the Hotspots (or Target Regions) page, click the name :
-
In the details page, click the link in the Processed File field:
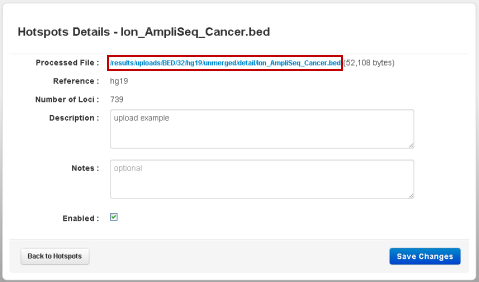
The Original File link in the Original Upload section also downloads the same file.
Delete a hotspots or target regions file
Follow these steps to delete a hotspots or a target regions file:
-
Go to the admin References tab and click either the Hotspots or Target Regions tab in the left navigation panel:

-
In the Hotspots (or Target Regions) page, click the name :

- In the details page, go to the Original Upload section and click the Delete button. If you are sure, click Yes in the confirmation popup.
BED file formats and examples
See BED File Formats and Examples .
Work with reference files
 Upload a New Reference File
Upload a New Reference File
 Delete a Reference Sequence
Delete a Reference Sequence
 Download an Ion Reference File
Download an Ion Reference File
 Details about the Ion hg19 Reference
Details about the Ion hg19 Reference
 Work with Obsolete Reference Sequences
Work with Obsolete Reference Sequences
Work with BED files
 Target Regions Files and Hotspot Files
Target Regions Files and Hotspot Files
 Manage Target Regions Files and Hotspot Files
Manage Target Regions Files and Hotspot Files
 BED File Formats and Examples
BED File Formats and Examples
 Manage DNA Barcodes and DNA Barcode Sets
Manage DNA Barcodes and DNA Barcode Sets
Work with reference library indices
 Update Reference Library Indices
Update Reference Library Indices
Work with test fragments


