References Management Guide
Torrent Suite Software space on Ion Community
References Management Guide TOC
Upload a New Reference File
As part of the standard analysis process, reads are aligned to a genomic reference, using the TMAP aligner that comes pre-installed on the Torrent Server. The alignments and some summary statistics based on the alignments are included in the Run Report Metrics Before Alignment of the Torrent Browser Analysis Report Guide .
For a new genome sequence, use the Admin > References tab to add the new reference genome. (These reference sequences are also displayed on the Ion S5, Ion PGM, or Ion Proton Sequencer when you load a sample.)
|
|
Internet Explorer 6.0 and earlier are not compatible with this tool. Please download the latest version of Internet Explorer (up to version 9), Firefox, or Chrome . |
Prerequistes
The following are prerequisites to uploading a new reference file:
-
Create a FASTA format reference sequence file (on your client machine).
- Prepare a descriptive name for the genome.
- Prepare the short name for the genome.
- Prepare a version for the genome.
- Know the number of reads to randomly sample for alignment.
- Prepare a regions of interest file or hotspots file (on your client machine).
Upload the reference
You upload reference files with the Admin gear menu References tab:
Follow these steps to upload a reference genome:
-
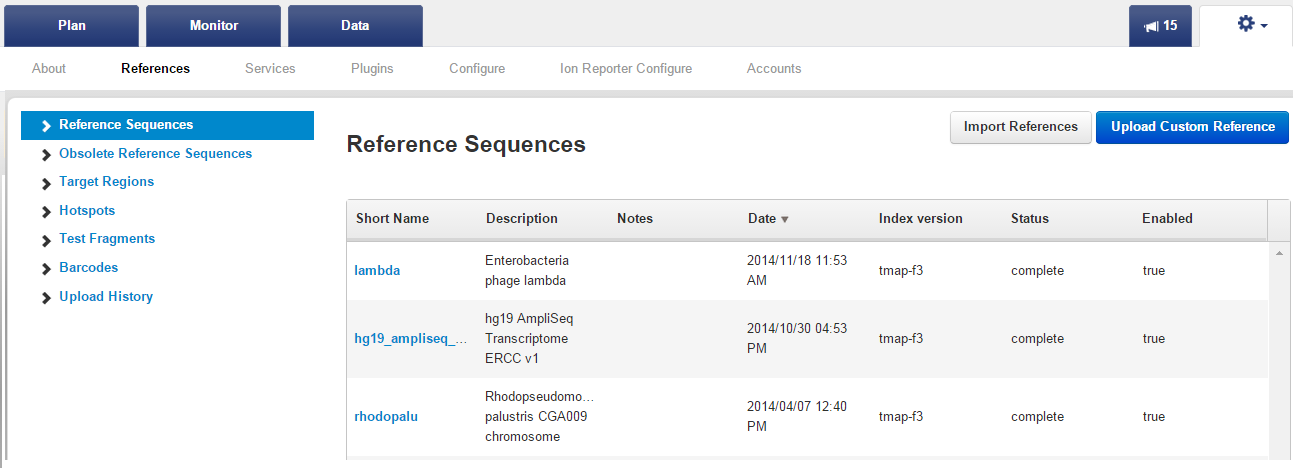
On the Admin > References tab, in the References Sequences section, click the
Upload a Custom Reference
button:
-
Fill out the Add New Reference Genome page. Required fields are noted on the page.
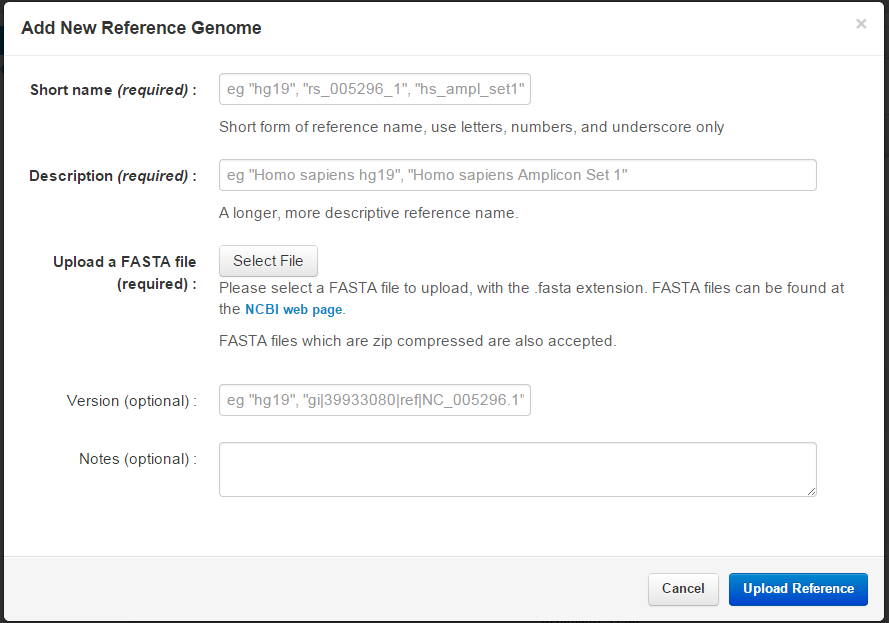
Field
Description
Descriptive name
[required]This entry may be any text string. The description usually includes the genus-species, version, and other identifying information. The description entered here is displayed in various report output, and is listed in the Reference Sequences section of the Admin > References tab.
Short name
[required] A shortened form of the genome name, the short form of the genome name may be any alphanumeric character and the underscore (_) character. The name should not match any existing references installed in the
/results/referenceLibrary/<index_type>/<genome_shortname>/directory, including previous unsuccessful attempts at creating reference sequences. Undesired sequences can be removed. Deletion allows the short name to be used for a new genome.Genome version
[required] Enter any string for the genome version number. The accession number, if there is one, is a good choice. The version entered here is displayed in various report outputs.
Notes
[optional] Use this field to record any notes about the reference genome
-
Click the
Select File
button and browse to the genome file (on your local machine).
-
Click the
Upload file and create reference
button.
-
Wait while the genome is uploaded.
After the reference is created, you can optionally add target regions BED files and hotspots BED or VCF files to the reference. See
Manage Target Regions Files and Hotspot Files
.
Error Handling
If you uploaded an invalidly formatted FASTA file, the following error displays when you attempt to view the reference sequence associated with the file:
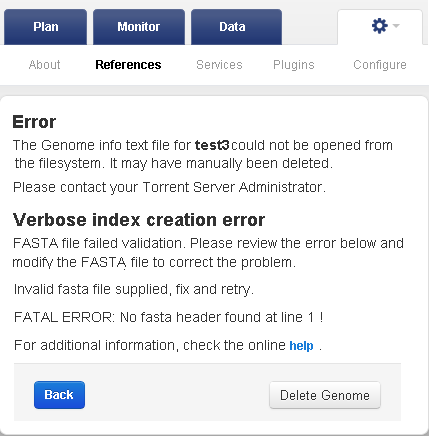
To recover from the error:
- Delete the existing reference sequence entry.
- Identify and correct formatting errors in the FASTA file.
- Retry Upload the reference .
Work with reference files
 Upload a New Reference File
Upload a New Reference File
 Delete a Reference Sequence
Delete a Reference Sequence
 Download an Ion Reference File
Download an Ion Reference File
 Details about the Ion hg19 Reference
Details about the Ion hg19 Reference
 Work with Obsolete Reference Sequences
Work with Obsolete Reference Sequences
Work with BED files
 Target Regions Files and Hotspot Files
Target Regions Files and Hotspot Files
 Manage Target Regions Files and Hotspot Files
Manage Target Regions Files and Hotspot Files
 BED File Formats and Examples
BED File Formats and Examples
 Manage DNA Barcodes and DNA Barcode Sets
Manage DNA Barcodes and DNA Barcode Sets
Work with reference library indices
 Update Reference Library Indices
Update Reference Library Indices
Work with test fragments


