The BaseCaller and Barcode Classification
Torrent Suite Software space on Ion Community
Overview of BaseCaller and Barcode Classification
This page discusses BaseCaller operations in general and issues around BaseCaller parameters, barcode classification, and filtering and trimming.
The settings of BaseCaller parameters control barcode classification as well as filtering and trimming.
About barcodes
Barcodes are short base sequences that during library preparation are placed between the library key and the read. The barcode sequences provide a mechanism to distinguish and identify reads from different samples during data analysis .
The use of barcodes allows multiple samples to be sequenced together on one chip during a sequencing run, and still have the run's read data be analyzed separately afterward as distinct samples.
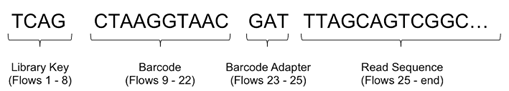
This diagram shows the placement of the barcode sequence, as well as the library key and adapters, with the read sequence (which is labeled "Template Bases"). The key is on the 5' end.

This example shows the location of the barcode sequence in both base space and flow space, using barcode IonPress_001 as an example:
Analysis pipeline overview
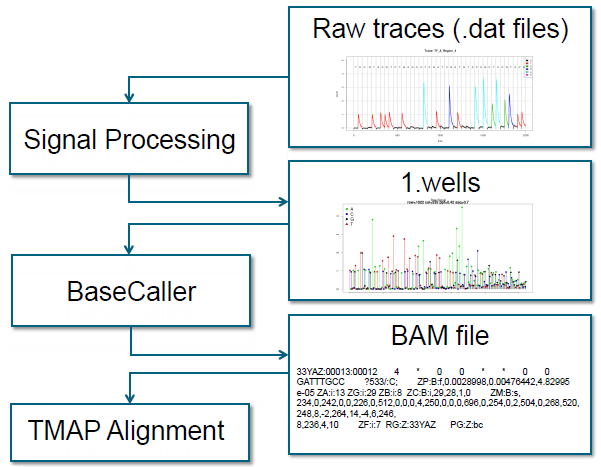
The beginning steps of the Torrent Suite Software analysis pipeline are shown below:
Steps:
- The sequencing instrument generates DAT files of electrical signals' raw traces.
- The signal processing step converts the raw traces into a single number per flow per well, in the 1.wells file.
- The BaseCaller converts the 1.wells file information into a sequence of bases and writes the sequence into an unaligned BAM file.
- The BAM file is passed to TMAP for alignment.
The signal processing step also marks several types of low-quality reads:
- Polyclonal reads (reads with two template beads instead of one)
- Reads with high signal processing residual (indicating an ambiguous signal value)
- Reads that do not contain a valid library key
The signal processing step marks these problematic reads but does not remove them.
Overview of BaseCaller functionality
In addition to creating a sequence of bases from the 1.wells file information, the BaseCaller module also performs read filtering and read trimming.
Notes on read filtering:
- Filters out low-quality reads that were marked during signal processing.
- Filters out reads that fail basecalling filters.
- Filtered out reads do not appear in the BAM file. The BaseCaller keeps counts of these reads but there is no record of specific reads that are filtered out.
Notes on read trimming:
- Removes certain bases from the read for quality reasons.
- The read appears in the BAM file.
-
The removed bases do not appear in the
BAM
file.
These are the steps performed in the BaseCaller:
- Remove low-quality reads that were marked during the signal processing step.
-
Do base calling:
- From the signal values, create the sequence of bases.
- Estimate the base quality value for each base.
-
Do barcode classification:
- Assign each read to a barcode.
- Trim the barcode sequence away.
-
Trim extra bases at the 5' end. Controlled by
--extra-trim-left(default is 0, meaning no extra trimming). -
Filter out reads that are too short.
Controlled by
--min-read-lengthand-- trim- min-read-len. -
Filter out reads that do not have the correct library key.
Can be turned off by
--keypass-filter. - Trim the P1 adapter (at the 3' end).
-
Perform quality trimming. Affect
ed by
--trim-qual-window-sizeand-- trim-qual-cutoff.
Notes about quality trimming:
- The purpose of quality trimming is to identify where quality issues begin at the end of a read. We try to identify when bases fall below a quality threshold and trim both those bases and a bit before those bases.
-
The parameter
--trim-qual-window-sizesets the window size for quality trimming. The algorithm slides through the sequence of bases and, each time the window shifts, computes the mean Base QV value for all bases in the window. -
If the
mean Base QV value for all bases in the window falls below a threshold (set the by parameter
--trim-qual-cutoff, default 16), then we trim all bases from the center of the window at that time to the 5' end.
Notes about barcode classification and barcode filtering
Barcode classification determines which barcode group a read is assigned to. Barcode classification is done for each read immediately after base calling.
Barcode filtering determines if a specific barcode is included in the run report or is filtered out. Barcode filtering works on the barcode groups as a whole
Other BaseCaller and barcode classification pages
Other pages related to barcode classification:
- BaseCaller Parameters (including changes in the 4.2 release)
- Troubleshooting Barcode Classification Issues
- Custom Barcode Design
- Filtering and Trimming Tech Note
 Introduction
Introduction
 Realign Run to Different Reference Genome
Realign Run to Different Reference Genome
 Reanalyze with a Different DNA Barcode Set
Reanalyze with a Different DNA Barcode Set
 Use DNA Barcodes with the Ion Torrent™ Sequencers
Use DNA Barcodes with the Ion Torrent™ Sequencers
 Overview of the BaseCaller and Barcode Classification
Overview of the BaseCaller and Barcode Classification
 Troubleshooting Barcode Classification Issues
Troubleshooting Barcode Classification Issues
 Custom Barcode Design
Custom Barcode Design
 Scan Your Sequencing Kit
Scan Your Sequencing Kit
 Handle a Failed Analysis Run
Handle a Failed Analysis Run
 Determine the Fault Cause
Determine the Fault Cause
 Restart a Run
Restart a Run
 Terminate an Analysis Run
Terminate an Analysis Run
 Work with Files
Work with Files
 Work with the Database
Work with the Database
 Change the Report Name
Change the Report Name
 Change the Run Date
Change the Run Date
 Add or Change an Ion PGM™ or Ion Proton™ Instrument
Add or Change an Ion PGM™ or Ion Proton™ Instrument
 Change Your Torrent Browser Password
Change Your Torrent Browser Password

