Torrent Suite SoftwareAdministration Guide
Torrent Suite Software space on Ion Community
Customer Support Archive
This page describes the contents of a Customer Support Archive and how you can generate one. The Customer Support Archive is a ZIP file of log files and other technical data about your Torrent Suite Software and analysis runs. This data is useful for our support team to diagnose issues with your Torrent Suite Software. A Customer Support Archive can only be generated by a request in your Torrent Browser an archive is not generated automatically or forwarded automatically to Ion Torrent.
Generate a Customer Support Archive
You can generate a Customer Support Archive from a run report in the Torrent Browser Data > Completed Runs & Reports tab. The Support tab appears in this tab ribbon (after the run metrics and before the plugin detailed output):
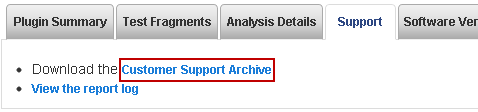
Click the Support tab and then the Customer Support Archive download link. Wait while the file is downloaded. The zipped archive file can be 15 or 20 MB or more.
Customer Support Archive contents
The tables in this section describe the files included in a Customer Support Archive. Files for optional modules (such as recalibration) only appear if the optional module is run.
In the top level directory:
| File | Description |
|---|---|
| alignment.log | L og of the final TMAP alignment process |
| < RunName >_< AnalysisReportName >.alignment.summary | Text format summary of sample alignment final results |
| alignment.summary |
Text format summary of sample alignment final results (same as the file <
RunName
>_<
AnalysisReportName
>.alignment.summary, but with a predictable file name)
|
| backupPDF.pdf |
PDF file of the analysis report and plugin results (similar to the output of the
Download as PDF
button on a run report)
|
| Controller | LiveView log of user activity on the sequencing instrument |
| debug | Log from datacollect, the background data acquisition module |
| DefaultTFs.conf | List of known Test Fragment sequences and their bases |
| drmaa_stderr_block.txt | Analysis pipeline error log for the block being executed by Sun Grid Engine |
| drmaa_stdout.txt | Log of events after primary analysis |
| drmaa_stdout_block.txt | Analysis pipeline output log for the block being executed by Sun Grid Engine |
| explog.txt | Initial runs settings needed for Torrent Browser analysis when being exported from instrument |
| explog_final.txt | Final runs settings needed for Torrent Browser analysis when being exported from instrument |
| InitLog.txt | Instrument auto pH log |
| InitValsW2.txt | pH log of the W2 solution |
| InitValsW3.txt | pH log of the W3 solution |
| RawInit.txt | Contains initialization data output |
| sysinfo.txt | Torrent Browser system software settings |
| TF.alignment.summary | Summary of test fragment alignment results in text file |
| uploadStatus |
Log of metrics being uploaded to the
Torrent Brows
er
|
| version.txt |
Torrent Suite software versions used for the analysis report
|
In the
basecaller_results
directory:
| File | Description |
|---|---|
| basecaller.log | Log file for the basecaller analysis module |
| datasets_basecaller.json | A JSON-format file of the settings needed for basecaller to analyze the sample data |
| datasets_pipeline.json | A JSON-format file of the settings needed by the pipeline to run the basecallermodule |
| datasets_tf.json | A JSON-format file of the settings needed for basecaller to analyze the Test Fragments |
| < RunName >_< AnalysisReportName >.quality.summary | A quality summary of basecaller unaligned reads/bases after filtering and trimming |
| quality.summary | Same as above, but with a predictable file name |
| TFStats.json |
A JSON-format file of
Test Fragments results statistics
|
In the
basecaller_results/recalibration
directory:
| File | Description |
|---|---|
| alignment.log |
L
og of the TMAP alignment process during base recalibration
|
| alignmentQC_out.txt | Log file from the TMAP analysis module |
In the
basecaller_results/unfiltered.trimmed
directory:
| File | Description |
|---|---|
| alignment.log |
L
og of the TMAP alignment process based on unfiltered and trimmed reads
|
| < RunName >_< AnalysisReportName >. alignment .summary | Text format summary of sample alignment results for unfiltered and trimmed reads |
| alignment.summary |
Text format summary of sample alignment results for
unfiltered and trimmed reads
(same as above, but with a predictable file name)
|
| datasets_basecaller.json |
A JSON-format file of the settings
needed for basecaller to analyze the sample data,
when generating the raw BAM file
|
| < RunName >_< AnalysisReportName >.quality.summary |
The
basecaller unfiltered and trimmed
reads/bases
quality summary
|
| quality.summary |
The
basecaller unfiltered and trimmed
reads/bases
quality summary (same as above, but with a predictable file name)
|
In the
basecaller_results/unfiltered.untrimmed
directory:
| File | Description |
|---|---|
| alignment.log |
L
og of the TMAP alignment process based on unfiltered and trimmed reads
|
| < RunName >_< AnalysisReportName >. alignment .summary | Text format summary of sample alignment results for unfiltered and untrimmed reads |
| alignment.summary |
Text format summary of sample alignment results for
unfiltered and untrimmed reads
(same as above, but with a predictable file name)
|
| datasets_basecaller.json |
A JSON-format file of the settings
needed for basecaller to analyze the sample data, when generating the raw BAM file
|
| < RunName >_< AnalysisReportName >.quality.summary |
The
basecaller unfiltered and untrimmed
reads/bases
quality summary
|
| quality.summary |
The
basecaller unfiltered and untrimmed
reads/bases
quality summary (same as above, but with a predictable file name)
|
In the
sigproc_results
directory:
| File | Description |
|---|---|
|
analysis.bfmask.stats |
Analysis statistics of wells in the bead find stag e (the bfmask is a set of bit flags for each well, indicating the contents of each well ) |
| avgNukeTrace_ATCG.txt | ATCG key signal measurements |
|
avgNukeTrace_
TCAG
.txt |
TCAG key signal measurements |
| bfmask.stats | Summary statistics of wells in the bead find stag e |
| processParameters.txt | Parameter settings for analysis signal processing |
| separator.bftraces.txt | Matrix data to separate between live wells and empty wells during bead find phase |
| separator.trace.txt | Matrix data to separate between live wells and empty wells |
| sigproc.log | Log file for the analysis module |
In the
sigproc_results/dcOffset
directory:
| File | Description |
|---|---|
| dcOffset.txt | background model parameter values of dcOffset |
In the
sigproc_results/NucStep
directory:
The files in this folder contain background model parameter values based on the location of the well in the chip .
| File |
|---|
| NucStep_frametime.txt |
| NucStep_inlet_head.txt |
| NucStep_inlet_empty.txt |
| NucStep_inlet_empty_sd.txt |
| NucStep_inlet_step.txt |
| NucStep_middle_head.txt |
| NucStep_ middle _empty.txt |
| NucStep_ middle _empty_sd.txt |
| NucStep_ middle _step.txt |
| NucStep_outlet_head.txt |
| NucStep_ outlet _empty.txt |
| NucStep_outlet_empty_sd.txt |
| NucStep_outlet_step.txt |
 Torrent Suite™ Software Administration Guide
Torrent Suite™ Software Administration Guide
 Deploy Your System
Deploy Your System
 Prepare Your Site
Prepare Your Site
 Install the Server
Install the Server
 Network Connectivity
Network Connectivity
 Update Torrent Suite™ Software
Update Torrent Suite™ Software
 Install Analysis Plugins
Install Analysis Plugins
 Configure Torrent Suite™ Software
Configure Torrent Suite™ Software
 Verify Functionality
Verify Functionality
 Manage Sequencer Settings from the Torrent Browser
Manage Sequencer Settings from the Torrent Browser
 Monitor Free Disk Space
Monitor Free Disk Space
 Back Up and Restore Data
Back Up and Restore Data
 Mount a USB Drive
Mount a USB Drive
 Boot Into Single-User Mode
Boot Into Single-User Mode
 Install and Use a UPS
Install and Use a UPS
 Axeda® Remote System Monitoring (RSM)
Axeda® Remote System Monitoring (RSM)
 Configure Chips
Configure Chips
 Configure Experiments
Configure Experiments
 Configure Global Configs
Configure Global Configs
 Configure Users
Configure Users
 Approve User Account Requests
Approve User Account Requests
 Configure Basecaller Default Parameters
Configure Basecaller Default Parameters
 Upgrade Your Torrent Server Ubuntu® Software
Upgrade Your Torrent Server Ubuntu® Software
 View Network Settings
View Network Settings
 Shutdown Server
Shutdown Server
 Use a Torrent VM to Evaluate a New Torrent Suite™ Software Release
Use a Torrent VM to Evaluate a New Torrent Suite™ Software Release
 Update Server
Update Server
 Update the Ion OneTouch™ Device
Update the Ion OneTouch™ Device
 Manage your SGE Cluster with QMON
Manage your SGE Cluster with QMON
 View System Support Diagnostics
View System Support Diagnostics
 View Instrument Diagnostics
View Instrument Diagnostics

