Torrent Browser Analysis Report Guide
Torrent Suite Software space on Ion Community
ERCC Analysis Plugin
This section describes the ERCC Analysis plugin. This plugin helps with analyses that use ERCC RNA Spike-in Controls. The plugin enables you to quickly determine whether or not the ERCC results indicate a problem with either the library preparation or the sequencing instrument run.
Enable the ERCC Analysis plugin
A plugin must be enabled before it can run. Your Torrent Suite Software administrator may have already enabled ERCC Analysis plugin and then the plugin appears in the run report Plugin Summary Select Plugin to Run list.
Follow these steps if you need to enable the plugin:
-
Scroll to the top of the Torrent Browser and click
Plugins
in the gear menu on the right:
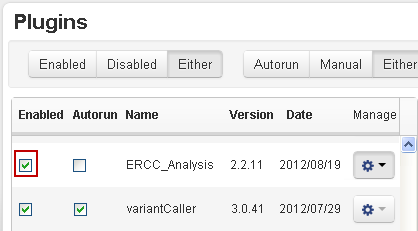
- If the ERCC_Analysis plugin does not appear on your plugin page, click the Name column to sort by name and scroll to the plugin. In the ERCC_Analysis row, click the Enable column checkbox:
Manually run the ERCC Analysis plugin
Follow these steps to manually launch the ERCC_Analysis plugin. You can use the default minimum R-squared value or enter you own value.
-
Open the run report for the analysis. Scroll down to the Plugin Summary section.
-
Click
Select plugin to run
to open the plugin list:
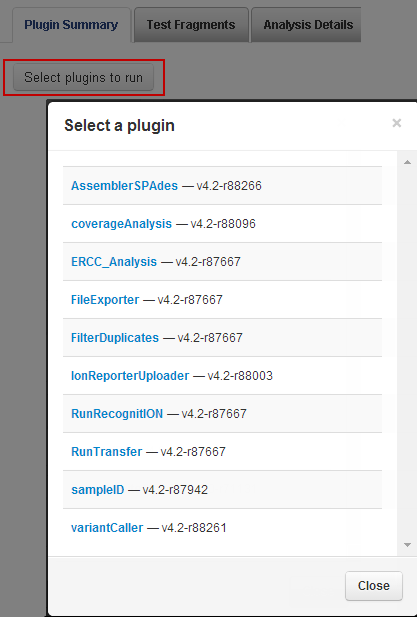
-
Click
ERCC_Analysis
. (The version number on your system might be different from the version shown here.)
-
A popup window with a minimum acceptable R-squared value appears. You can use the default value or enter your own value.The value should be in the range from 0 to 1.
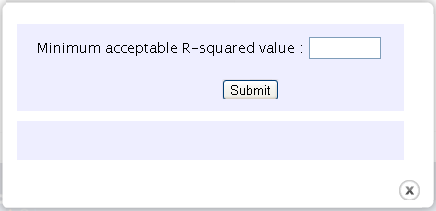
-
Click Submit to start the plugin.
Configure a template to run the ERCC plugin
If you configure a template or planned run to execute the ERCC Analysis plugin, and your experiment uses the Ion Total RNA-Seq Kit V2, then on the wizard Kits page, you must make a Barcode Set selection. Select either one of the following in the Barcode Set menu:
- IonXpressRNA Select this if your experiment uses this kit.
- RNA_Barcode_None Select this if your experiment does not use a barcode kit. This selection is required for the correct trimming.
The following wizard image shows the two barcode kit selections. (Make only one selection per run.)

Plugin run times
For analysis runs with total reads under 1,000,000, the plugin normally takes 2-3 minutes to run (on supported hardware). For larger runs, the plugin takes approximately an additional 1-2 minutes per million total reads. For example, a run with 5 million reads may take 10-15 minutes. These run times are offered only as a guideline. If your Torrent Suite Software is busy with other processing, plugin run times are longer.
After the ERCC analysis is completed, you can view the analysis results.
View analysis results
Plugin analysis results appear in the plguin summary area:

After the status of the ERCC plugin has changed to Completed, click the ERCC_Analysis.html link in the Plugin Summary section to open the ERCC Report and view analysis results:
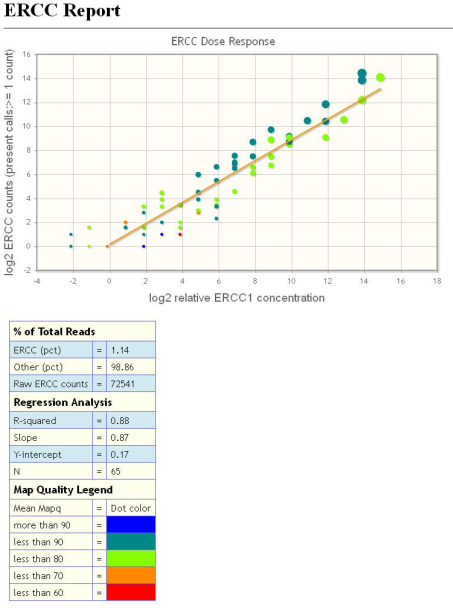
Interpret the data
The ERCC Report screen (shown on page 5) displays the ERCC Dose Response plot. The points are color-coded, based on mapping quality. There is also a trendline, based on the parameters shown in tabular form to the right of the graph.
The y-axis of the plot is the log (base 2) of the raw counts found for the transcript in question. The x-axis is also logarithmic, but represents the known relative concentration of the ERCC transcripts. Ideally, the points all fall on a straight line.
More realistically, in the good case, the raw counts and relative concentration should at least correlate with a high R-squared (for example, 0.9 or higher). The table to the right of the plot (shown on page 5) shows the R-squared value found for this plot, as well as the Slope, Y-intercept, and N (number of transcripts found) values. Although there are 92 transcripts in the ERCC mix, it is not expected that all 92 will be detected. The number of transcripts detected depends on the sequencing depth.
View transcript details
If you want to look at the details regarding a particular transcript, there are two methods you can use:
-
Hover your mouse-cursor over a point on the ERCC Dose Response plot to display a popup window that shows details about that transcript (the name, reads, and coverage plots).
If several points are very close together on the plot and it is difficult to hover over the point you are interested in, you can zoom in on the plot to more easily distinguish points:
-
- Use your mouse to draw a box around the point of interest and magnify it.
- To zoom out to the full view of the ERCC Dose Response plot, either doubleclick the plot, or click the Reset Zoom button.
-
Scroll to the particular transcript, and click the [+] next to the transcript name.
This method shows the same information, plus a few additional pieces. See Definitions if the meaning of any of these pieces is unclear.
Definitions
This section defines terms used in the plugin output.
- Coverage Depth The minimum and maximum number of reads covering bases in the transcript. If coverage is 100%, the minimum value will be > 0.
- Coverage The number of base positions covered by at least one read.
- Start Sites The number of base positions that are the start site for a read.
- Unique Start Sites The number of start sites that have only one read starting at the site.
- Coverage CV Coefficient of Variation for coverage = average coverage / stddev coverage for the entire transcript.
( Optional ) Configure the ERCC Analysis plugin
You can optionally change the R-squared value to set a default value for the summary report screen:
1. If the window showing the minimum acceptable R-squared value is open, close it. Then scroll to the top of the Torrent Browser and click Plugins in the gear menu on the top right:

2. If the ERCC_Analysis plugin does not appear on your plugin page, click the Name column to sort by name and scroll to the plugin. In the ERCC_Analysis row, click the Manage column gear menu, then select Configure .
3. The ERCC Plugin Configuration screen opens:
Enter a value between 0 and 1 as your minimum acceptable R-squared value (a lower value is indicated by a red light in the summary report). Then click Submit .
The value you enter on the ERCC Plugin Configuration screen is used when the plugin is auto-run and when a user manually launches the plugin without entering a value. Users can override this value on a per-run basis when they manually launch the plugin.
View plugin error information
If the ERCC_Analysis plugin status changes to Error after you click the ERCC_Analysis.html link, then something went wrong during the running of the plugin. In this case, look at the error log:
-
Return to the Plugin Summary, then click the log file icon to see the error log:

-
Scroll to the bottom of the log to see error messages:
An Error status for the ERCC_Analysis plugin should be a rare event and indicates that the ERCC_Analysis plugin itself failed to run. A plugin error does not indicate that the ERCC_Analysis results are bad.
ERCC resources
The External RNA Controls Consortium (ERCC) is hosted by the U.S. National Institute of Standards and Technology. The ERCC Analysis plugin is for experiments that use ERCC RNA Spike-In Control Mixes, set of RNA controls derived from the ERCC plasmid reference library.
For information on ERCC RNA Spike-In Control Mixes, please refer to the ERCC RNA Spike-In Control Mixes User Guide (Pub no. 4455352).
For more information on ERCC analysis, refer to the following resources:
Figure 2, Analysis of ERCC read counts, in
Sensitivity of RNA-Seq using Ion semiconductor sequencing a comparison to microarrays and qPCR
The Ion Torrent white paper
Methods, tools, and pipelines for analysis of Ion PGM Sequencer miRNA and gene expression data
The information on the
ERCC ExFold RNA Spike-In Mix
product page.
 Torrent Browser Analysis Report Guide
Torrent Browser Analysis Report Guide
 Run Report Metrics
Run Report Metrics
 Run Metrics Overview
Run Metrics Overview
 Run Report Metrics Before Alignment
Run Report Metrics Before Alignment
 Run Report Metrics on Aligned Reads
Run Report Metrics on Aligned Reads
 Barcode Reports
Barcode Reports
 Test Fragment Report
Test Fragment Report
 Report Information
Report Information
 Output Files
Output Files
 Plugin Summary
Plugin Summary
 Assembler SPAdes Plugin
Assembler SPAdes Plugin
 Coverage Analysis Plugin
Coverage Analysis Plugin
 ERCC Analysis Plugin
ERCC Analysis Plugin
 FileExporter Plugin
FileExporter Plugin
 FilterDuplicates Plugin
FilterDuplicates Plugin
 IonReporterUploader Plugin
IonReporterUploader Plugin
See
 The Ion Reporter™ Software Integration Guide
The Ion Reporter™ Software Integration Guide
 Run RecognitION Plugin
Run RecognitION Plugin
 SampleID Plugin
SampleID Plugin
 TorrentSuiteCloud Plugin
TorrentSuiteCloud Plugin
 Torrent Variant Caller Plugin
Torrent Variant Caller Plugin
 Torrent Variant Caller Parameters
Torrent Variant Caller Parameters
 Example Torrent Variant Caller Parameter File
Example Torrent Variant Caller Parameter File
 Torrent Variant Caller Output
Torrent Variant Caller Output
 The Command-Line Torrent Variant Caller
The Command-Line Torrent Variant Caller
 Ion Reporter™ Software Features Related to Variant Calling
Ion Reporter™ Software Features Related to Variant Calling
 Integration with TaqMan® and PCR
Integration with TaqMan® and PCR

