Torrent Browser Analysis Report Guide
Torrent Suite Software space on Ion Community
Torrent Variant Caller Plugin
- Introduction
- Templates
- Supported Ion AmpliSeq panels
- Parameter Settings defaults
- Variant Caller parameters
- Upload your custom parameter values
- Input files
- Run the Torrent Variant Caller plugin
- Torrent Variant Caller plugin manual launch for custom configuration per barcode
- Torrent Variant Caller plugin output
- Rerun the variantCaller plugin
- Integration with other Thermo Fisher sites
- Send feedback
Introduction
The Torrent Variant Caller (TVC) plugin calls single-nucleotide polymorphisms (SNPs), multi-nucleotide polymorphisms (MNPs), insertions, and deletions in a sample across a reference or within a targeted subset of that reference.
This plugin provides optimized pre-set parameters for many experiment types but is also very customizable. After you find a parameter combination that works well on your data and that has the balance of specificity and sensitivity that you want, you can save that parameter set and reuse it over and over in your research. This is supported on both manual launches of the plugin and in automatic launches through the run plan template wizard.
Parameters
TVC provides several ways of handling its parameter options:
- You can select one of TVC default pre-set parameter groups. TVC provides these defaults that are optimized for several experiment types.
- You can start with one of TVC default pre-set parameter groups and then make your own customizations in the TVC UI.
- You can import parameter settings that are optimized for fixed panels and community panels in ampliseq.com. (Optimized parameter sets for custom designs are not supported in this release.)
- You can download the parameters used in a TVC run and then either customize those parameters or reuse them in future TVC runs.
TVC's default parameters setting groups are organized according to these attributes:
- Variant frequency Somatic settings are optimized to detect low frequency variants. Germ-line settings are optimized for high frequency settings.
- Sequencing instrument The Ion S5, Ion PGM , or the Ion Proton sequencer. Parameter defaults are different for Ion Proton data than for Ion PGM and Ion S5 data.
- Stringency High stringency settings are optimized to minimize false positives. Low stringency settings minimize false negatives.
- TargetSeq Two sets of defaults are optimized for Ion TargetSeq data.
See Parameter Settings defaults and Variant Caller parameters for more information about TVC's parameters.
Reference
The Reference Genome field names the reference used in the original Torrent Suite Software. The reference cannot be changed.
Library Type
The Library Type selection does not change or customize TVC's parameter settings. When the Library Type is set to AmpliSeq, the Trim Reads option is available. Trimming is recommended for Ion AmpliSeq data to remove the adapters from the reads.
Targeted Regions and HotSpot Regions menus
If you select Targeted Regions or HotSpot Regions files fromtheir dropdown menus, and they are applied to your analysis:
- Targeted Regions Analysis is restricted to only the regions of interest that you specify in this file.
- Hotspots Variant Caller output files include these positions whether or not a variant is called, and include evidence for a variant and the filtering thresholds that disqualified a variant candidate.
See also Input files .
Templates
Be aware that the TVC plugin is not selected in the templates shipped with Torrent Suite Software. Select the plugin when you create a run plan or a copy of a shipped template.
Templates that you download from ampliseq.com do have the TVC plugin selected.
Supported Ion AmpliSeq panels
The TVC plugin supports the various panels in the Ion AmpliSeq family of sequencing kits, including the following:
- Ion AmpliSeq BRCA1 and BRCA2 research panel
- Ion AmpliSeq Colon and Lung Cancer research panel
- Ion AmpliSeq CFTR research panel
See Create a Template with Ion AmpliSeq.com Import for steps to import settings optimized for the above panels into your TVC run.
The following table lists the TVC parameter options that are pre-defined and optimized for ampliseq.com panels or Ion TargetSeq data:
| Panel or application | TVC Pre-set parameter defaults |
|---|---|
|
Ion AmpliSeq Exome |
Germline - Proton - Low Stringency |
|
CCP PGM |
Somatic - PGM - Low Stringency |
|
CCP Proton |
Somatic - Proton - Low Stringency |
|
CHP2 (HSM2) |
Somatic - PGM - Low Stringency |
|
CHv1 |
Somatic - PGM - Low Stringency |
|
IDP |
Germline - PGM - Low Stringency |
|
Ion TargetSeq data |
Germline - Proton TargetSeq - Low Stringency |
Parameter Settings defaults
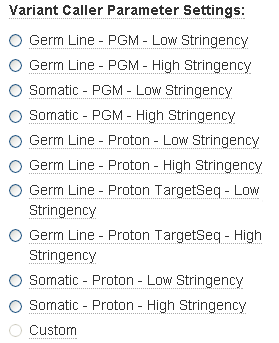
The Torrent Variant Caller parameter settings change according to your Variant Caller configuration radio button selection. Data from Ion PGM and Ion Proton Sequencers require different default settings. Select settings that are appropriate to both your sequencing instrument and your experiment:
-
Germ-Line - Low Stringency Optimized for high frequency variants and minimal false negative calls.
-
Germ-Line - High Stringency Optimized for high frequency variants and minimal false positive calls.
-
Somatic - Low Stringency Optimized for low frequency variant detection with minimal false negative calls.
-
Somatic - High Stringency Optimized for low frequency variant detection with minimal false positive calls.
-
Germ-Line - TargetSeq Low Stringency Optimized for high frequency variants and minimal false nega tive calls. (Ion Proton data only)
-
Germ-Line - TargetSeq High Stringency Optimized for high frequency variants and minimal false positive calls. (Ion Proton data only)
-
Custom Settings that you customize. (You cannot select this radio button. This button is enabled if you change a parameter value.)
About the use of Variant Caller Parameter Settings radio buttons
First select the appropriate Variant Caller Parameter Settings radio button. Your radio button selection loads the correct set of default parameters for that type of run. If you want to customize parameters further, change parameter values in the main settings area. Advanced users can also click the Show Advanced Settings button to change values in the advanced settings.
These notes apply to the Variant Caller Parameter Settings and advanced settings selections:
-
If you do customize settings in the advanced settings area, your changes are overwritten if you select a different Variant Caller Parameter Settings radio button (or again click on the same radio button).
-
If you make changes in the advanced settings and later want to reset these parameters to their default values, again click your Variant Caller Parameter Settings radio button selection.
Variant Caller parameters
Individual TVC parameters are described in Torrent Variant Caller Parameters .
In general, you can safely customize parameters for SNP calling. For indel calling, changes to the parameters tend to have a significant effect in the number of indels called. With indels, the tradeoff between sensitivity and specificity becomes too large.
Parameters are categorized as main settings, which are intended for general use, and advanced settings, which allow additional customization of the variant calling algorithm but are intended for advanced users only.
Note: To download the parameters used in a completed TVC run, see Parameters File in Buttons for downloads and other actions .
Upload your custom parameter values
Use the Upload Custom Settings Choose File button to upload your set of custom parameter settings:
You can use this mechanism for the following:
-
To quickly apply your own settings to all your TVC plugin runs
-
To know that your parameters are consistent (for instance, that a parameter change is not inadvertently forgotten in the UI)
-
To apply a file of settings shared by others
The parameters file must be in JSON format. For an example of this type of file, see Example Torrent Variant Caller Parameter File .
After upload, the UI reflects the parameter values from your uploaded file. You can still make additional changes in the UI.
Follow these steps to upload a parameters file for your TVC plugin run:
-
Have the
JSON
file to be uploaded on your local machine. You can optionally edit values in the file before uploading.
-
In the TVC plugin launch page, click the
Choose File
button under Upload Custom Parameter Settings:

- Browse to your parameters file and click OK .
The optimized parameters are imported into your run and are reflected in the parameter table on the launch page.
Input files
This section describes input files that you provide for the TVC plugin.
Both a target regions file and a hotspots file must be associated with a reference before you use them with the TVC plugin. You upload these files to a specific reference, such as hg19, in the admin References page. See Manage Target Regions Files and Hotspot Files .
Target regions file
A target regions file controls the sequencing and downstream analysis of a targeted resequencing run in this way: sequencing is restricted to specified chromosome regions that appear in the regions of interest file. (In contrast, a whole genome analysis sequences every position that corresponds to the reference genome.)
The regions of interest file must be a Browser Extensible Data (BED) file, which is a tab-separated file format. For format information, see Manage Target Regions Files and Hotspot Files .
Hotspots file
A hotspots file contains a list of positions on the genome and when configured in a workflow affects the analysis results. For each position during variant calling:
-
Evidence for a variant is examined at that position (without regard to the hotspots positions) and a call is made.
-
Then the hotspots positions are examined. At each position listed in the file, if a variant is not already called, then one of the following variant calls is added:
- REF Homozygous reference
- NO_CALL A variant is not called at this position (for instance, because of lack of coverage)
- The filtering metrics for each position are reported in the output VCF file, including for NO_CALLs.
The hotspots file must be in BED or VCF format. For format information, see Manage Target Regions Files and Hotspot Files .
By default the variantCaller plugin calls variant candidates at hotspot positions with more sensitivity than candidates at other positions. You can customize certain variantCaller parameters separately for hotspot candidates.
Run the Torrent Variant Caller plugin
There are two ways to run the Torrent Variant Caller plugin: automatically, by preconfiguring the plugin to run as soon as primary analysis has completed, or manually, allowing you to run the plugin at any time from a completed run report.
See also Variant Caller parameters .
Torrent Variant Caller plugin manual launch for custom configuration per barcode
New in Torrent Suite Software v5.0, you can configure individual barcodes in a run to be processed with their own reference genome, target regions file, hotspots file and TVC parameters. Please note, that this functionality is only available via manual launch of TVC after the run and not available from the run planning stage.
- Select a completed run that you would like to reanalyze with TVC.
-
Click
Report Actions
and
Select Plugins to Run
.
- Select variantCaller . The Torrent Variant Caller 5.0 plugin configuration screen appears.
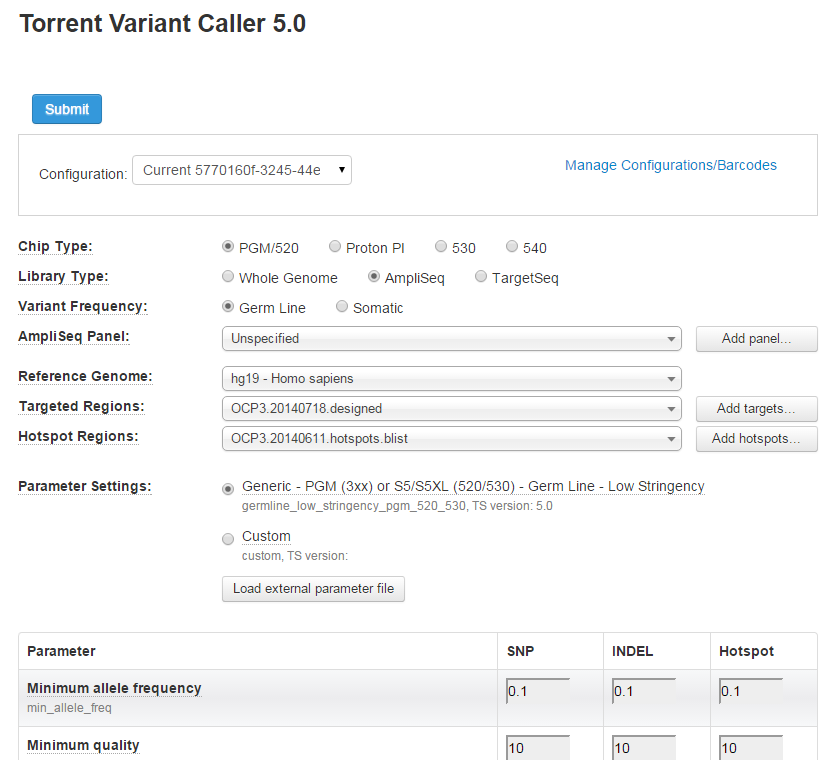
Apply TVC Settings to all Barcodes
To apply the same or different settings, select from the Configuration drop-down menu.
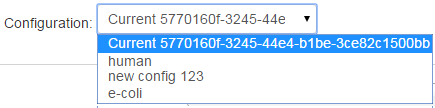
Modify and Apply TVC Settings for All or Select Barcodes
To customize your TVC settings, click the Manage Configurations/Barcodes link. The Configuration tab allows you to Edit, Delete or Add a configuration. The Setup tab allows you to apply settings to individual barcodes.
Apply TVC changes per barcode
-
Click the Setup tab and modify settings per individual barcodes.
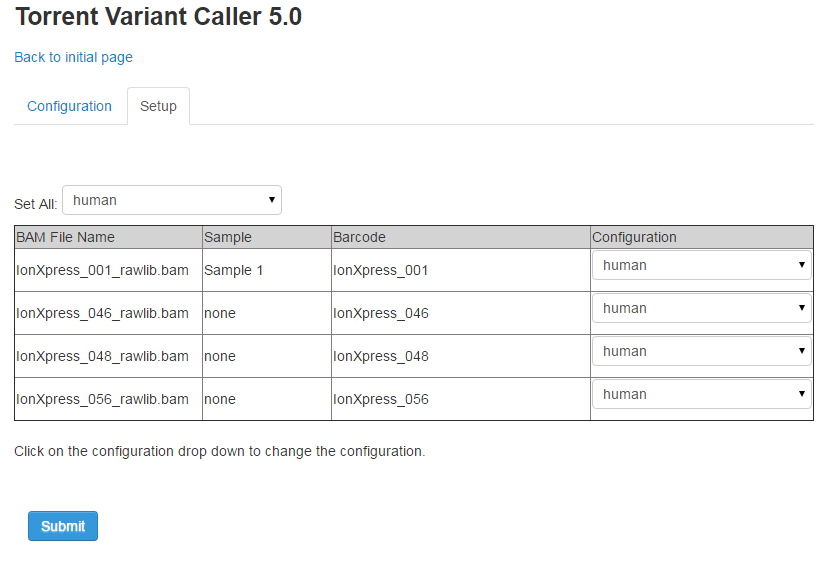
-
When finished making changes, click Submit. The TVC plugin reruns and applies the changes you made.
Add, Edit, and Delete Configurations
- On the Configuration tab, you can Add new or Edit/Delete existing configurations.
-
Click the
Add
button to add new. Name the configuration and select your settings.
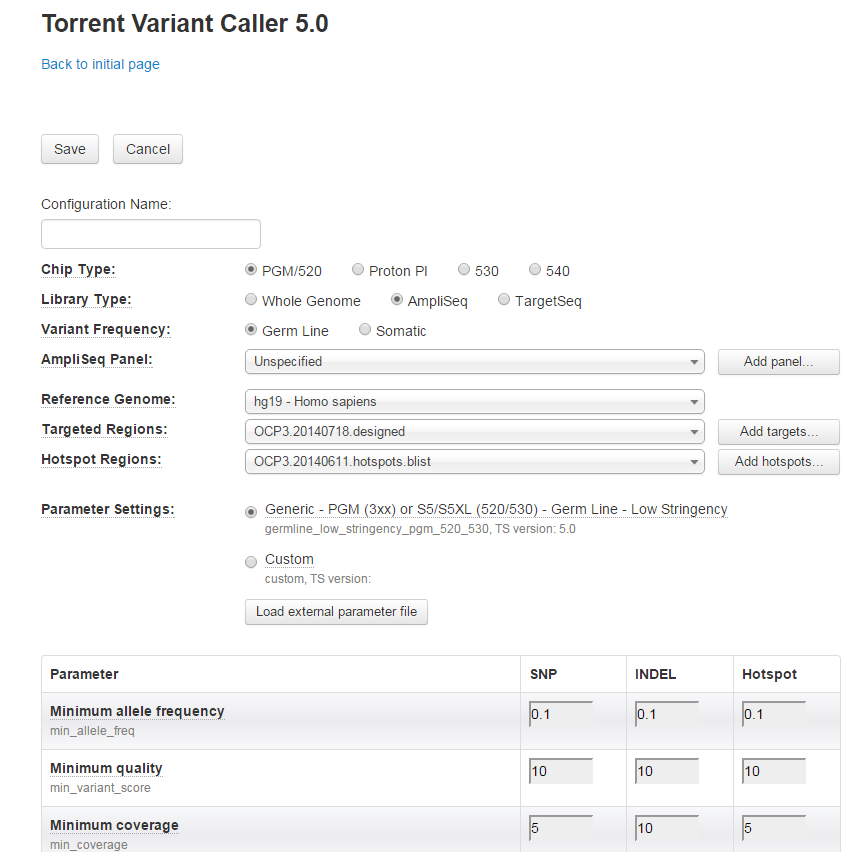
-
On the Edit screen, you can modify Chip and Library types, variant frequency, reference genome, targeted and hotspots regions and parameter settings.
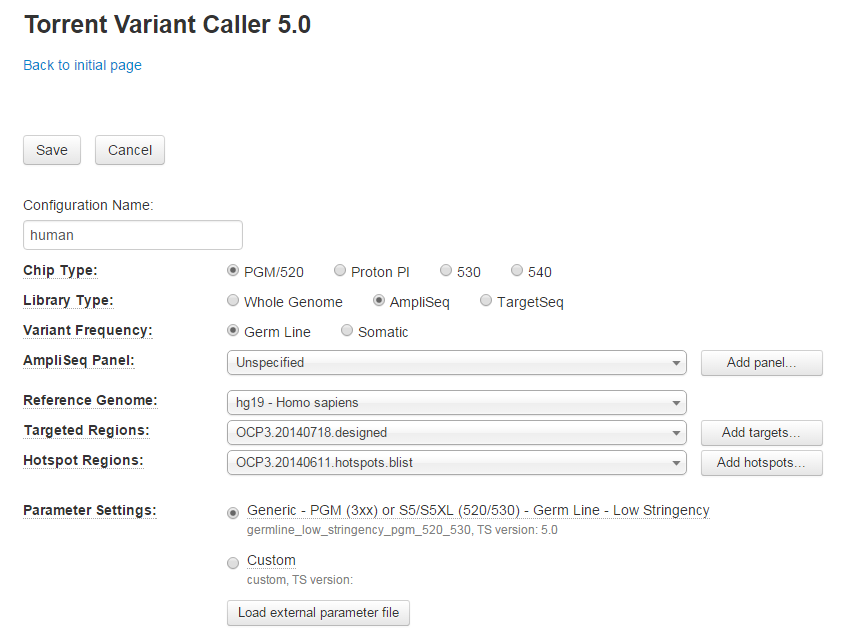
- At the bottom of the screen, you can click the Show Advanced Settings button and further adjust variant detection and alignment parameters.
- Save your new or modified configurations and then apply them to all or select barcodes.
- Click Submit to rerun TVC.
Configure the Torrent Variant Caller in a template or run plan
Use the run plan template wizard to have the TVC plugin run automatically after the Torrent Suite analysis completes.
Before following these steps, read the Introduction , Supported panels , Parameter Settings defaults , and Variant Caller parameters sections for information about the options and customizations for the plugin.
Note : The TVC plugin run uses the same target regions file and hotspots file as the main Torrent Suite Software analysis (if those files are present in the main analysis). Through the wizard there is no facility in the TVC configuration to change the target regions file or hotspots file. You can use a different target regions file and hotspots file with a manual TVC launch from a completed run report.
When you select the plugin chevron in the template or run plan wizard and enable the variantCaller checkbox, a Configuration link appears next to the
variantCaller listing
:
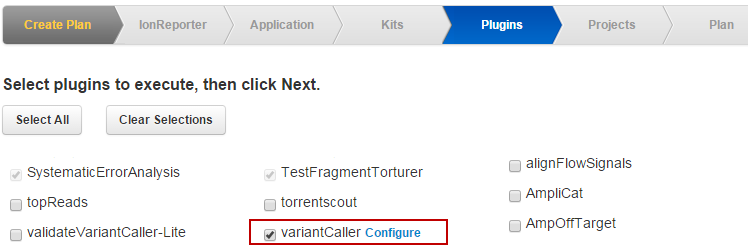
Link the Configure link to open the variantCaller configuration popup:
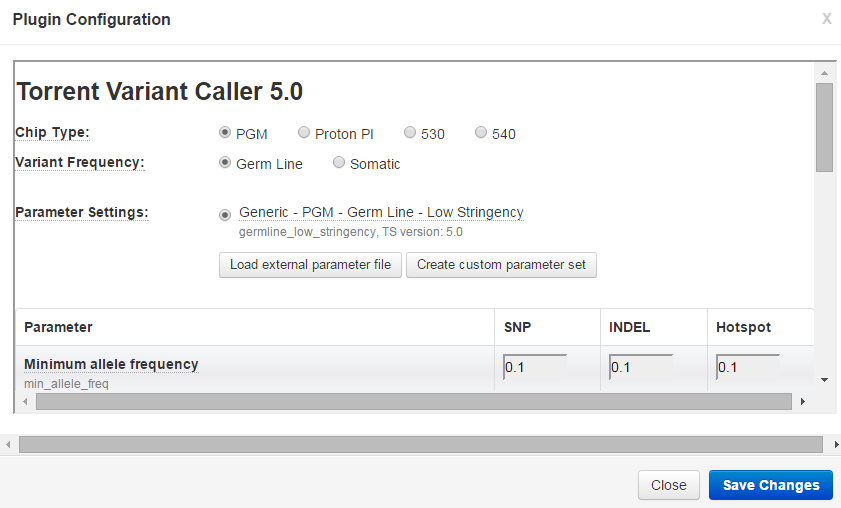
- Make your changes to the parameter values.
- Advanced users can also click the Show Advanced Settings button and customize those parameters.
- Be sure to click the Save Changes button before leaving this page.
Be sure to click the Save Plugin Settings button before you click Close . Your changes are lost if you do no use the Save Changes button.
You can later return to the Variant Caller configuration page by clicking the Configure button next to variantCaller in the Plugin chevron.
Note: Changes to parameters can dramatically affect the behavior and sensitivity of the Variant Caller. Parameter changes are not recommended if you are new to the Variant Caller plugin.
Run the Torrent Variant Caller manually
The TVC plugin supports multiple run analysis. The plugin can analyze a BAM file generated from Combine Alignment on multiple reports in a project. Combine Alignment creates a new run report (in the same project). Y ou can open the new combined run report and use the Select plugins to run button to launch the TVC plugin.
To run the Torrent Variant Caller plugin manually, perform the following steps:
- In the Torrent Browser, select a run report on the Data > Completed Runs & Reports page or on a Data > Projects > projectname page.
-
Click
Report Actions
>
Select plugins to run
.
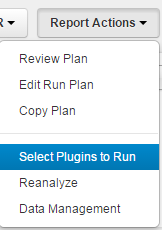
-
In the
list of plugins, click on
variantCaller
.
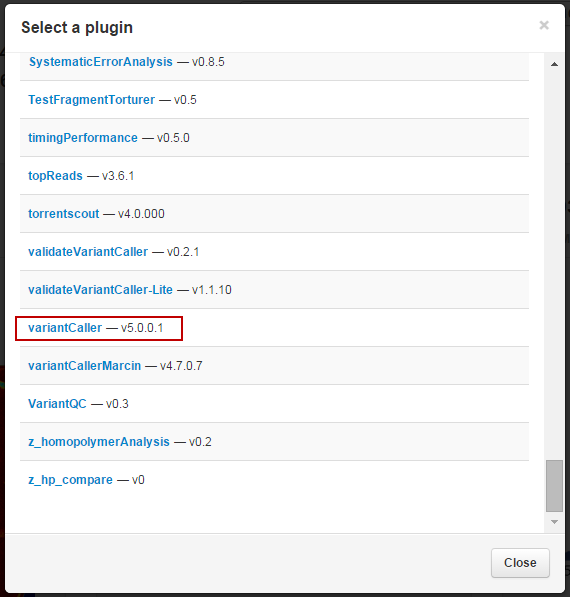
The Torrent Variant Caller Plugin interface appears:
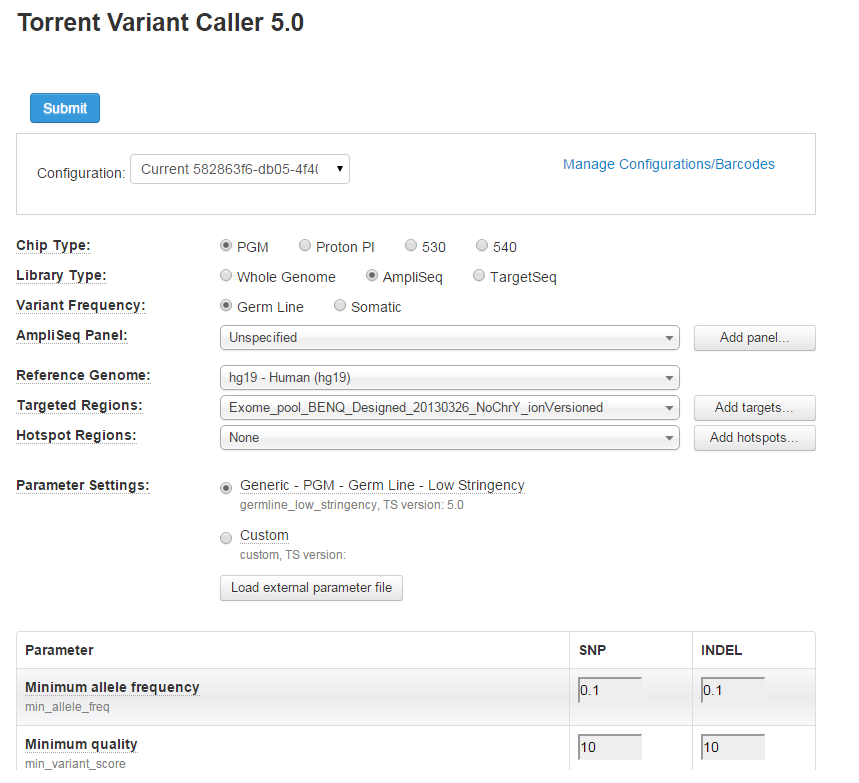
These options are described in the
Introduction
,
Input files
,
Supported panels
,
Parameter Settings defaults
, and
Variant Caller parameters
VCopts
sections.
When you are satisfied with your selections, click Submit . Your variant caller analysis is queued for execution.
Torrent Variant Caller plugin output
The Torrent Variant Caller plugin output includes the following reports and sections:
Run report plugin summary
Variant Caller Report
Variant Caller Report s ummary section
Variant Caller Report Variant Calls table
See Torrent Variant Caller Output for descriptions of the reports and tables.
Run report plugin summary
The run report contains a short summary of plugin output. These summaries appear below the run report metrics and above the Output Files section.
To jump to the plugin results:
Click the Plugin Summary jump near the top of the run report:
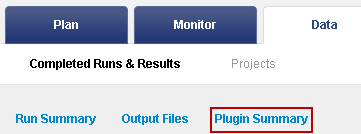
Click the link to go to the TVC results summary:

The browser jumps to the variantCaller summary.
The variantCaller summary area is slightly different for barcoded and non-barcoded runs. In both cases, the summary section includes the following:
Information about the analysis type, targeted regions and hotspot files, and variantCaller parameter settings.
The total number of variants called.
The variantCaller.html link to the results page.
Download links:
- The zipped VCF file of variant calls.
- The Zipped VCF index file (required for IGV).
- The results in a tab-separated file.
- The genome VCF file
Barcoded variantCaller summary area
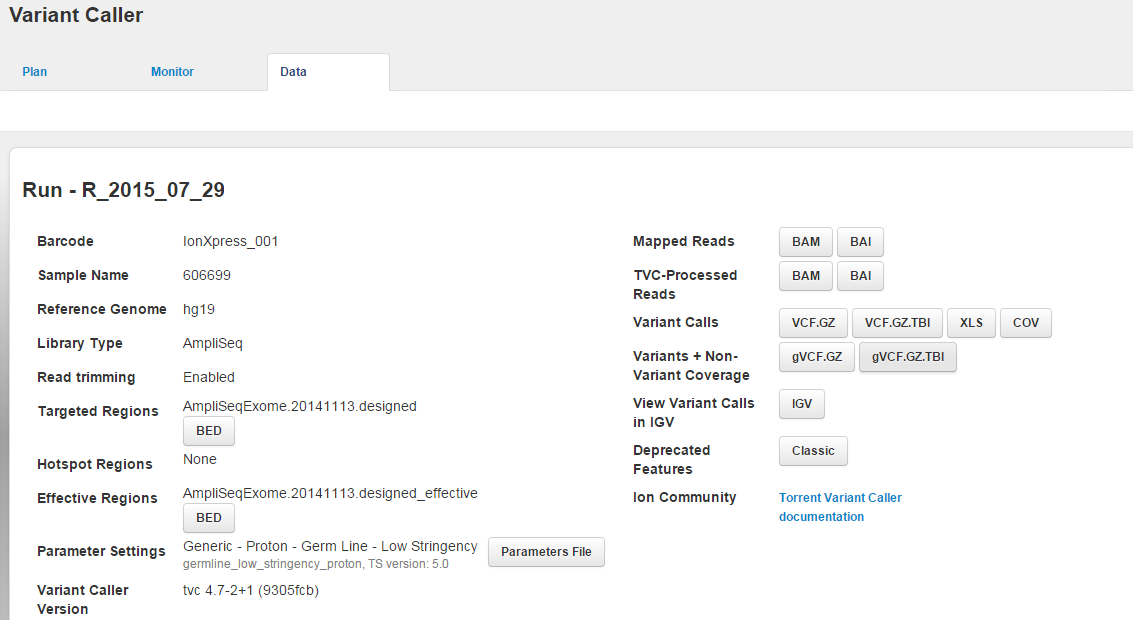
For a barcoded run:
When the run contains multiple barcodes, the variantCaller.html link opens a listing of the barcodes.
Links to a separate results page for each barcode.
A link to download all results in one zipped file.
Non-barcoded variantCaller summary area
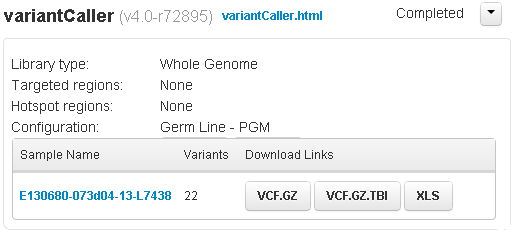
For a non-barcoded run, the sample name is listed. This link and the variantCaller.html link open the same results page.
View plugin log file
To view the log file for a plugin run, in the plugin summary area, click the menu arrow next to the plugin status:
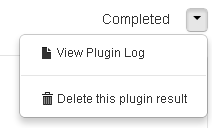
Delete plugin results
To delete the results for a plugin run, in the plugin summary area, click the menu arrow next to the plugin status:

This action removes the plugin's output files from the file system and removes the Torrent Browser plugin results page.
Download files, other actions
|
Field |
Button |
Description |
|---|---|---|
|
Targeted regions |
BED |
Downloads the input targeted regions BED file (if any). |
|
Hotspot regions |
BED |
Downloads the input hotspots BED file (if any). |
|
Parameters Settings |
Parameters File |
Downloads a JSON text file of the TVC parameter values used on this run. Note: You can edit this file and later upload it to set your custom parameters in subsequent runs. For an example of this type of file, see Example Torrent Variant Caller Parameter File . |
|
Mapped Reads |
BAM, BAI |
Downloads t he BAM file (and its index) of mapped reads. This file is input to TVC. |
|
Variant Calls |
VCF.GZ,
|
Downloads files of the variants calls:
VCF.GZ,
VCF.GZ.TBI: Zipped VCF file and its tabix index file
gVCF.GZ: Reports positions on the genome where no variants were found. |
|
Open Variants Calls in IGV |
IGV |
Link to open the results variants in the Integrated Genomic Browser (IGV). |
|
Deprecated Features |
Classic |
Opens the plugin results page in the previous format. |
| Ion Community | Torrent Variant Caller documentation | Opens to the Torrent Variant Caller documentation page on the Ion Community (login is required). |
Export to file
This option exports your variant calls to a tab-separated file.
The exported file is named
subtable.xls
and has the same columns as the Variant Calls table (including columns for all three display options: View Allele Annotations,
View Coverage Metrics, and View Quality Metrics).
Click the left column checkboxes to select your variants, then click the Export Selected button:
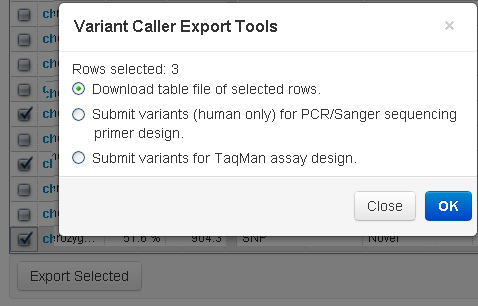
Rerun the variantCaller plugin
You can rerun the variantCaller plugin from the results page:
Scroll to the Adjust Parameters area at bottom of the results page, and click
Show Filter Settings
:

In the parameter listings,make your changes to the parameter settings (only main parameters are available):
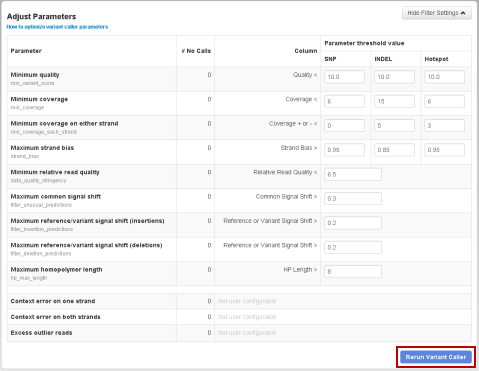
Click Rerun Variant Caller . The plugin is submitted for execution.
Integration with other Thermo Fisher sites
See Torrent Variant Caller Integration with TaqMan and PCR for the following:
To select variants to search for pre-designed primers for PCR and Sanger sequencing
To select variants to submit a search on the TaqMan Assay Search webpage
Send feedback
Use the Send Feedback button, in the bottom right of the browser, to send your comments about the Torrent Variant Caller plugin:

The feedback app asks if your comments are about a specific part of the current page or if your comments are more general:
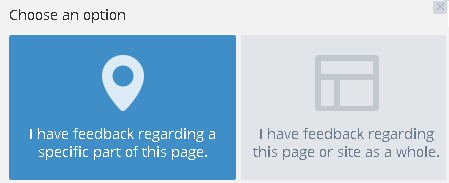
If you select the specific part of the current page option, you next click on that feature in the page and then fill out a text form with your comments.
See
 Torrent Browser Analysis Report Guide
Torrent Browser Analysis Report Guide
 Run Report Metrics
Run Report Metrics
 Run Metrics Overview
Run Metrics Overview
 Run Report Metrics Before Alignment
Run Report Metrics Before Alignment
 Run Report Metrics on Aligned Reads
Run Report Metrics on Aligned Reads
 Barcode Reports
Barcode Reports
 Test Fragment Report
Test Fragment Report
 Report Information
Report Information
 Output Files
Output Files
 Plugin Summary
Plugin Summary
 Assembler SPAdes Plugin
Assembler SPAdes Plugin
 Coverage Analysis Plugin
Coverage Analysis Plugin
 ERCC Analysis Plugin
ERCC Analysis Plugin
 FileExporter Plugin
FileExporter Plugin
 FilterDuplicates Plugin
FilterDuplicates Plugin
 IonReporterUploader Plugin
IonReporterUploader Plugin
 The Ion Reporter™ Software Integration Guide
The Ion Reporter™ Software Integration Guide
 Run RecognitION Plugin
Run RecognitION Plugin
 SampleID Plugin
SampleID Plugin
 TorrentSuiteCloud Plugin
TorrentSuiteCloud Plugin
 Torrent Variant Caller Plugin
Torrent Variant Caller Plugin
 Torrent Variant Caller Parameters
Torrent Variant Caller Parameters
 Example Torrent Variant Caller Parameter File
Example Torrent Variant Caller Parameter File
 Torrent Variant Caller Output
Torrent Variant Caller Output
 The Command-Line Torrent Variant Caller
The Command-Line Torrent Variant Caller
 Ion Reporter™ Software Features Related to Variant Calling
Ion Reporter™ Software Features Related to Variant Calling
 Integration with TaqMan® and PCR
Integration with TaqMan® and PCR

