Torrent Browser Analysis Report Guide
Torrent Suite Software space on Ion Community
Assembler SPAdes Plugin
This plugin assembles reads into long sequences ( contigs ) and allows for basic level analysis, with metrics such as number of contigs, N50, and other analysis metrics. The plugin is ideal for genomes less than 50 megabases in size. The plugin assumes a haploid genome. For multiploid genomes, reads from different copies of a chromosome tend to assemble into different contigs.
Run the AssemblerSPAdes plugin on a completed run
Follow these steps to run the plugin and to review the plugin output report.
-
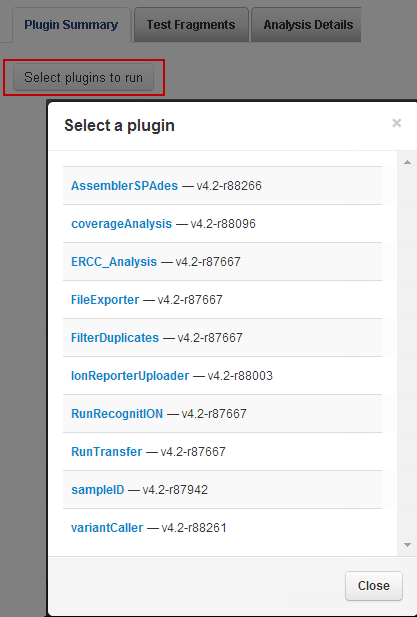
To run this plugin, in the report for your run, scroll down to the Plugin Summary section, and click
Select plugins to run
.
-
In
Select a plugin
, select
Assembler SPAdes
.Theplugin displays the plugin user interface:
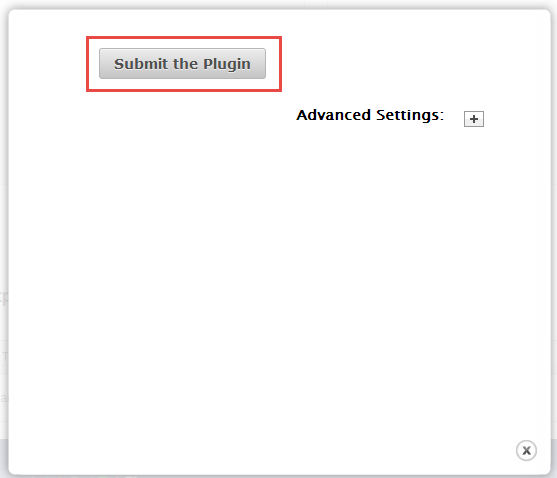
For most uses, you can take the defaults and click Submit the Plugin .
Advanced settings
This section describes the optional advanced settings. To open the advanced parameters fields, click the Advanced Settings + button.
These are the default advanced settings:
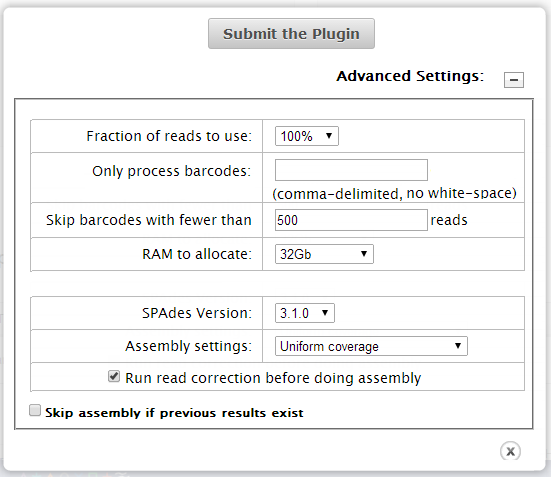
| Field | Description |
|---|---|
| Fraction of reads to use |
If less than 100%, reads are randomly sub-sampled. 100% is recommended the plugin automatically handles most changes in coverage. |
| Only process barcodes | By default the plugin processes all barcodes in the analysis and produces separate set of contigs for each barcode. To limit plugin analysis to only specific barcodes, list those barcodes here (separated by commas, no spaces). Example: |
| Skip barcodes with fewer than | Ignores barcodes whose number of reads do not meet this threshold. Intended to filter out barcode classification problems with noisy data. |
| RAM to allocate |
The plugin attempt to allocate the amount of RAM specified here. With larger amounts of memory, the plugin runs faster. With less memory, the plugin takes longer to complete. Note: The plugin crashes if the memory allocation fails. |
| SPAdes version | Select the version that you prefer. Select the default if you do not know. |
| Assembly settings |
Set this menu as follows:
|
| Run read correction before doing assembly | Recommended. |
| Skip assembly if previous results exist | Recommended. |
Custom assembly settings
The K and Mode fields are displayed when the Assembly settings menu is set to Custom:
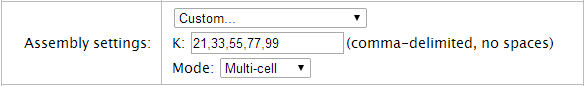
The K field
The SPAdes plugin is a De-Bruijin graph assembler. The plugin breaks reads in kmers, makes a connected graph, and traverses through that graph to produce contigs.
The K field determines the kmer size and how many kmers are used.
Notes about the kmers setting:
- Use a smaller kmer if your data contians many errors.
- If your data contains many repeats, use a larger kmer.
- Each additional kmer adds a fixed amount to the processing time (using 2 kmers takes twice as long as one kmer).
The Mode menu
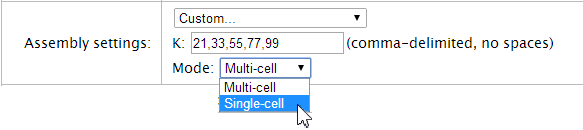
Select the Single-cell option for data with high (> 68%) GC content.
Select the Multi-cell option for data with average or low GC content.
Ouptut
The plugin output includes sections for downloads and statistics:
Downloads
- Assembled Contigs Download or open a FASTA file of the assembled sequences in the FASTA format.
- SPAdes log Download or open a text file of log messages from SPAdes. (For more information , see the SPAdes site bioinf.spbau.ru/spades .)
- QUAST repor t Download or open an HTML file of assembly statistics generated by QUAST. (For more information, see SPAdes documentation http://spades.bioinf.spbau.ru/release2.1.0/quality.htm .)
Assembly Statistics
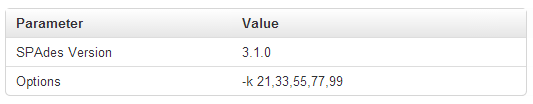
Parameter section
The Parameter section shows the version of SPAdes that generated this run and the set of kmers that was used:
Metric section
The Metric section shows the following information:
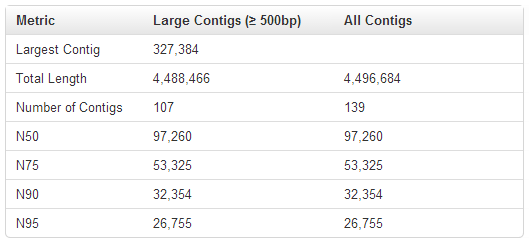
| Metric | Description |
|---|---|
| Largest Contig | Length of the longest assembled contig |
| Total Length | Total number of base pairs contained in all assembled contigs that are at least 500 bp in length. |
| Number of Contigs | N umber of assembled contigs that are at least 500 bp in length. |
| N50 | The contig length such that using longer or equal length contigs produces half (50%) the bases of the assembly. Usually there is no value that produces exactly 50%, so the more technical definition is the minimal length x such that using contigs of length at least x accounts for at least 50% of the total assembly length. |
| N75 | The contig length such that using longer or equal length contigs produces 75% of the bases of the assembly. |
| N90 | The contig length such that using longer or equal length contigs produces 90% of the bases of the assembly. |
| N95 | The contig length such that using longer or equal length contigs produces 95% of the bases of the assembly. |
 Torrent Browser Analysis Report Guide
Torrent Browser Analysis Report Guide
 Run Report Metrics
Run Report Metrics
 Run Metrics Overview
Run Metrics Overview
 Run Report Metrics Before Alignment
Run Report Metrics Before Alignment
 Run Report Metrics on Aligned Reads
Run Report Metrics on Aligned Reads
 Barcode Reports
Barcode Reports
 Test Fragment Report
Test Fragment Report
 Report Information
Report Information
 Output Files
Output Files
 Plugin Summary
Plugin Summary
 Assembler SPAdes Plugin
Assembler SPAdes Plugin
 Coverage Analysis Plugin
Coverage Analysis Plugin
 ERCC Analysis Plugin
ERCC Analysis Plugin
 FileExporter Plugin
FileExporter Plugin
 FilterDuplicates Plugin
FilterDuplicates Plugin
 IonReporterUploader Plugin
IonReporterUploader Plugin
See
 The Ion Reporter™ Software Integration Guide
The Ion Reporter™ Software Integration Guide
 Run RecognitION Plugin
Run RecognitION Plugin
 SampleID Plugin
SampleID Plugin
 TorrentSuiteCloud Plugin
TorrentSuiteCloud Plugin
 Torrent Variant Caller Plugin
Torrent Variant Caller Plugin
 Torrent Variant Caller Parameters
Torrent Variant Caller Parameters
 Example Torrent Variant Caller Parameter File
Example Torrent Variant Caller Parameter File
 Torrent Variant Caller Output
Torrent Variant Caller Output
 The Command-Line Torrent Variant Caller
The Command-Line Torrent Variant Caller
 Ion Reporter™ Software Features Related to Variant Calling
Ion Reporter™ Software Features Related to Variant Calling
 Integration with TaqMan® and PCR
Integration with TaqMan® and PCR

