Torrent Browser User Interface Guide
Torrent Suite Software space on Ion Community
Wizard KitsChevron
On the Kits wizard page, enter the following information about laboratory kits and other sequencing parameters:
- ( Optional )Sample preparation kit
- Library kit type, including the forward library key and the forward 3' adapter
- Templating kit type
- Sequence kit
- Number of flows
- Barcode set Required for barcoded runs
- Base calibration mode
- Control sequence Required for RNA runs
- Chip type Required
- Mark PCR Duplicates Not recommended for Ion AmpliSeq data
Chip type is now required. As with all fields, if you enter chip type in your templates, then it is automatically entered in your run plans.
Example Kits chevron:
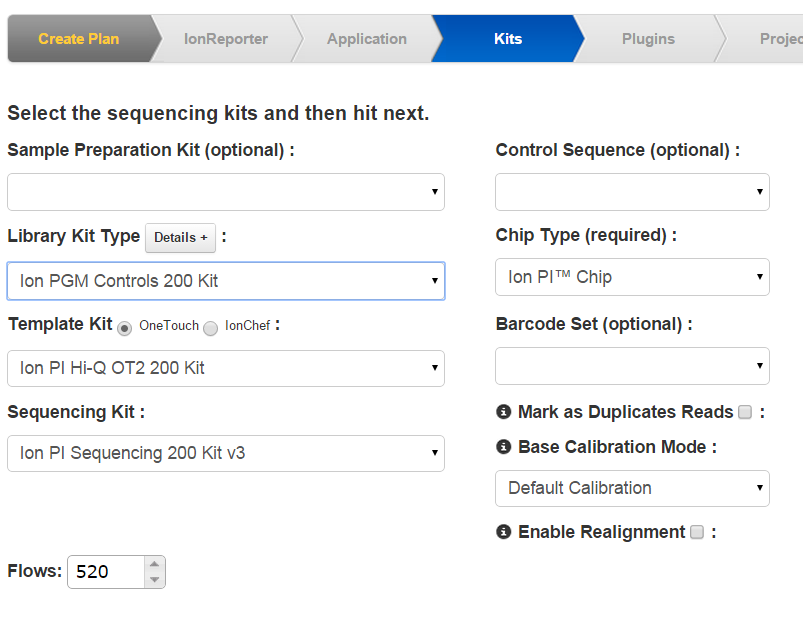
Note: The value entered for number of flows represents the maximum possible for a run using a planned run based on this template. Instrument conditions such as the availability of consumables might cause fewer flows to be completed.
If the Barcode Set menu is grayed out
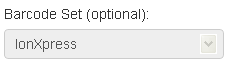
When you plan by sample set, the barcode set is specified in the Barcoding chevron. In that case, the menu is grayed out in the Kits chevron.
About the Base Calibration mode options
New in Torrent Suite 4.4, there is a base calibration mode drop-down menu that contains three options: Default Calibration, No Calibration and Enable Calibration Standard. (Previously, in Torrent Suite 4.2, you could choose to Enable Base Recalibration or not.) You can select the base calibration method during run planning and in the reanalysis menu.
Default Calibration - allows a random subset of wells to be used for base calibration. (This is equivalent to the default setting for Torrent Suite 4.2 and earlier, i.e., a checked Enable Base Recalibration check box).
No Calibration allows you to turn off base calibration. (This is equivalent to an uncheckedEnable Base Recalibration check box in TS 4.2 and earlier.)
Enable Calibration Standard allows wells belonging to the Calibration Standard to be selected as training subset.
The Calibration Standard is a small panel consisting of known sequence content with comprehensive and uniform representation of long homopolymers (up to 10-mers). The calibration standard can be spiked into Ion S5, Ion PGM, and Ion Proton runs as a quality control for higher homopolymer performance and as a known reference for base recalibration.Please note that this method of base calibration only works if calibration standard beads were spiked into the run.A summary of the number of calibration standard beads found can be viewed under the Calibration Report tab on the run page.
About the Mark as PCR Duplicates option
For some applications, duplicate reads coming from PCR cause problems in downstream analysis. The presence of duplicate reads may create the appearance of multiple independent reads supporting a particular interpretation, when some of the reads are in fact duplicates of each other with no additional evidence for the interpretation.
Torrent Suite Softwareuses an Ion-optimized approach that considers the read start and end positions by using both the 5' alignment start site and the flow in which the 3' adapter is detected. Duplicate reads are flagged in the BAM in a dedicated field. Use of the Torrent Suite Softwaremethod is recommended over other approaches which consider only the 5' alignment start site.
Marking duplicate reads is not appropriate for Ion AmpliSeq data, because many independent reads are expected to share the same 5' alignment position and 3' adapter flow as each other. Marking duplicates on Ion AmpliSeq data risks inappropriately flagging many reads that are in fact independent of one another.
Wizard pages:
 Torrent Browser User Interface Guide
Torrent Browser User Interface Guide
 The Login Page
The Login Page
 The Plan Tab
The Plan Tab
 Templates
Templates
 Planned Runs
Planned Runs
 Plan by Sample Set
Plan by Sample Set
 Create Samples and a Sample Set
Create Samples and a Sample Set
 Sample Attributes
Sample Attributes
 Template and Planned Run Wizard
Template and Planned Run Wizard
 Create Multiple Run Plans
Create Multiple Run Plans
 Create a Template with Ion AmpliSeq.com Import
Create a Template with Ion AmpliSeq.com Import
 The Monitor Tab
The Monitor Tab
 The Data Tab
The Data Tab
 Completed Runs and Reports Tab
Completed Runs and Reports Tab
 Work with Completed Runs
Work with Completed Runs
 Reanalyze a Completed Run
Reanalyze a Completed Run
 BaseCaller Parameters
BaseCaller Parameters
 TMAP Parameters
TMAP Parameters
 The Projects Listing Page
The Projects Listing Page
 Project Result Sets Page
Project Result Sets Page
 Compare Multiple Run Reports
Compare Multiple Run Reports
 CSV Metrics File Format
CSV Metrics File Format

